| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
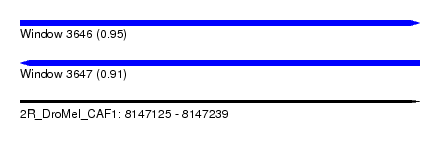
| Location | 8,147,125 – 8,147,239 |
| Length | 114 |
| Max. P | 0.948122 |

| Location | 8,147,125 – 8,147,239 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.54 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -24.44 |
| Energy contribution | -25.22 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8147125 114 + 20766785 UCAAAGGUUUAUAGCA---UGAUCUAUGCUGGGGAUUCCUUUCUUACCAGACACUUAUGCAGUUCGCUGAUCAGCGGGUGCUCCAGUUGCACAUUCGUUUGCUCCUGGAGCGCCCCA .............(((---(((....(((((((((......)))..)))).)).)))))).....(((....)))(((((((((((..(((........)))..))))))))))).. ( -42.30) >DroPse_CAF1 21364 99 + 1 GUAAAUC--ACUGAAA---U---------UACAGAG----UCAUUACCAGGUAUUUGUGCAGUUCACUGAUGAGCGGGUGCUCCAGUUUCACAUUUGUUUGUUCCUGAAGCGCCCCA ((..(((--(.((((.---.---------((((((.----.............))))))...)))).))))..))(((((((.(((....(((......)))..))).))))))).. ( -27.14) >DroSec_CAF1 33647 114 + 1 UCAAAGGUUUUUAACA---UGAUCUAUGCUGGGGAUUCUUUUCUUACGAGACACUUAUGCAGUUCGCUGAUCAGCGGGUGCUCCAGUUGCACAUUCGUUUGCUCCUGAAGCGCUCCA ....(((((.......---.)))))..((((((((......))))..............(((....)))..))))(((((((.(((..(((........)))..))).))))).)). ( -28.80) >DroSim_CAF1 24521 114 + 1 UCAAAGGUUUUUAACG---UGAUCUAUGCUGGGGAUUCCUUUCUUACCAGACACUUAUGCAGUUCGCUGAUCAGCGGGUGCUCCAGUUGCACAUUCGUUUGCUCCUGGAGCGCCCCA ................---(((((..(((((((((......)))..)))).)).....((.....)).)))))(.(((((((((((..(((........)))..)))))))))))). ( -39.30) >DroEre_CAF1 23370 117 + 1 UCAAAGAAUUAUAAAAUUUUGAUCUAUGCUAGGCAUUCCUUACUUACCAGACACUUAUGCAGUUCGCUUAUCAGCGGGUGCUCCAGUUGCACAUUCGUCUGCUCCUGGAGCGCCCCA .....(((((((((...((((......(..(((....)))..)....))))...)))))..))))(((....)))(((((((((((..(((........)))..))))))))))).. ( -33.20) >DroYak_CAF1 33740 107 + 1 ----------AAAACAUUUUGAUCUAUACUGGGCAUUCCUUACUUACCAGACACUUAUGCAGCUCGCUAAUCAGUGGGUGCUCCAGUUGCACAUUCGUCUGCUCCUGGAGCGCCCCA ----------.........((((.....((((..............))))........((.....))..))))(.(((((((((((..(((........)))..)))))))))))). ( -33.84) >consensus UCAAAGGUUUAUAACA___UGAUCUAUGCUGGGGAUUCCUUUCUUACCAGACACUUAUGCAGUUCGCUGAUCAGCGGGUGCUCCAGUUGCACAUUCGUUUGCUCCUGGAGCGCCCCA .............................((((((.....))))))...................(((....)))(((((((((((..(((........)))..))))))))))).. (-24.44 = -25.22 + 0.78)

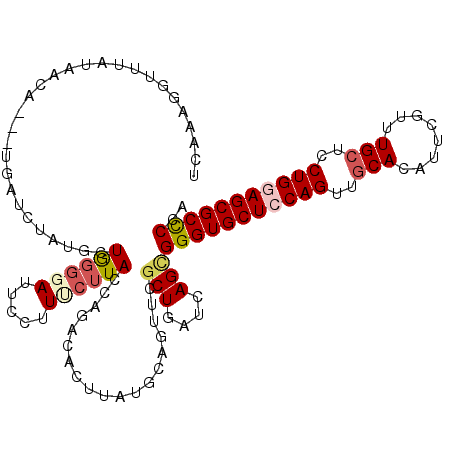

| Location | 8,147,125 – 8,147,239 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.54 |
| Mean single sequence MFE | -33.31 |
| Consensus MFE | -25.69 |
| Energy contribution | -26.75 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8147125 114 - 20766785 UGGGGCGCUCCAGGAGCAAACGAAUGUGCAACUGGAGCACCCGCUGAUCAGCGAACUGCAUAAGUGUCUGGUAAGAAAGGAAUCCCCAGCAUAGAUCA---UGCUAUAAACCUUUGA ..(((.(((((((..(((........)))..))))))).)))(((((((.((.....))....(((.((((..(........)..))))))).)))))---.))............. ( -40.10) >DroPse_CAF1 21364 99 - 1 UGGGGCGCUUCAGGAACAAACAAAUGUGAAACUGGAGCACCCGCUCAUCAGUGAACUGCACAAAUACCUGGUAAUGA----CUCUGUA---------A---UUUCAGU--GAUUUAC ..(((.(((((((..(((......)))....))))))).)))....((((.((((.((((.................----...))))---------.---.)))).)--))).... ( -25.05) >DroSec_CAF1 33647 114 - 1 UGGAGCGCUUCAGGAGCAAACGAAUGUGCAACUGGAGCACCCGCUGAUCAGCGAACUGCAUAAGUGUCUCGUAAGAAAAGAAUCCCCAGCAUAGAUCA---UGUUAAAAACCUUUGA .((((((((((((..(((........)))..)))))))...((((....))))....)).........((....))......)))..(((((.....)---))))............ ( -27.70) >DroSim_CAF1 24521 114 - 1 UGGGGCGCUCCAGGAGCAAACGAAUGUGCAACUGGAGCACCCGCUGAUCAGCGAACUGCAUAAGUGUCUGGUAAGAAAGGAAUCCCCAGCAUAGAUCA---CGUUAAAAACCUUUGA ..(((.(((((((..(((........)))..))))))).)))(((((((.((.....))....(((.((((..(........)..))))))).)))))---.))............. ( -37.60) >DroEre_CAF1 23370 117 - 1 UGGGGCGCUCCAGGAGCAGACGAAUGUGCAACUGGAGCACCCGCUGAUAAGCGAACUGCAUAAGUGUCUGGUAAGUAAGGAAUGCCUAGCAUAGAUCAAAAUUUUAUAAUUCUUUGA ..(((.(((((((..(((.(....).)))..))))))).)))(((....)))...........((((..((((.........))))..))))......................... ( -34.60) >DroYak_CAF1 33740 107 - 1 UGGGGCGCUCCAGGAGCAGACGAAUGUGCAACUGGAGCACCCACUGAUUAGCGAGCUGCAUAAGUGUCUGGUAAGUAAGGAAUGCCCAGUAUAGAUCAAAAUGUUUU---------- ..(((((((((((..(((.(....).)))..))))))).((.(((.(((((...(((.....)))..))))).)))..))...))))....................---------- ( -34.80) >consensus UGGGGCGCUCCAGGAGCAAACGAAUGUGCAACUGGAGCACCCGCUGAUCAGCGAACUGCAUAAGUGUCUGGUAAGAAAGGAAUCCCCAGCAUAGAUCA___UGUUAUAAACCUUUGA ..(((.(((((((..(((........)))..))))))).)))(((((((((...(((.....)))..)))))......((....))))))........................... (-25.69 = -26.75 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:59 2006