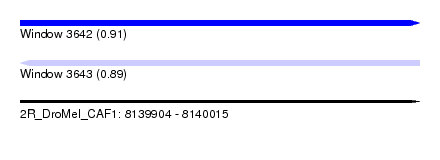
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,139,904 – 8,140,015 |
| Length | 111 |
| Max. P | 0.914976 |

| Location | 8,139,904 – 8,140,015 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -22.63 |
| Energy contribution | -23.47 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
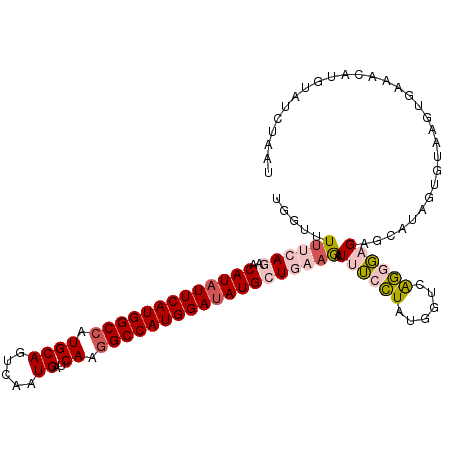
>2R_DroMel_CAF1 8139904 111 + 20766785 UGGUUUUUCAGAACAUAUUCAUGGCCAUGCAGUCAAUGAUCAAGGCCAUGGAUAUGCUGAAGAUUUCCUACGGUCAGGGAGAGCAUAGUGUAAGUGAAACAUUGAUCUAAU .((.(((((((..(((((((((((((.((((.....))..)).))))))))))))))))))))...))...((((((......(((.......))).....)))))).... ( -34.60) >DroVir_CAF1 37272 111 + 1 UGGUCUACCAAAACAUAUUCAUGGCCAUGCAGUCCAUGAUCAAGGCCAUGGACAUGCUCAAAAUAGAAUAUGGUGGUGCAGAGAUUCGUGUAAGCACUAAACAUAAUUAUG .(((((((((...(((((((........((((((((((........))))))).)))........))))))).))))....))))).(((....))).............. ( -29.89) >DroGri_CAF1 18944 111 + 1 UGGUGUUCCAAAACAUAUUCAUGGCCAUGCAGUCCAUGAUCAAAGCCAUGGAUAUGCUCAAAAUAGAAUAUGGUGGUCCAGAGAUUCAAGUAAGCUCUACAUGUAUUUAAG (((....(((...(((((((........((((((((((........))))))).)))........))))))).))).))).((((.((.(((.....))).)).))))... ( -27.09) >DroSim_CAF1 17097 111 + 1 UGGUUUUUCAGAACAUAUUCAUGGCCAUGCAGUCAAUGAUCAAGGCCAUGGAUAUGCUGAAGAUUUCCUAUGGUCAGGGAGAGCAUAGUGUAAGUGAAUCAUUUACCCAAU (((.(((((((..(((((((((((((.((((.....))..)).))))))))))))))))))))....(((((.((.....)).))))).(((((((...)))))))))).. ( -38.40) >DroEre_CAF1 17112 111 + 1 UGGUUUUUCAGAACAUAUUCAUGGCCAUGCAGUCAAUGAUCAAGGCCAUGGAUAUGCUGAAGAUUUCCUAUGGUCAGGGAGAACAUAGUGUAAGUGAAUCACGGACCUCAU .((.(((((((..(((((((((((((.((((.....))..)).))))))))))))))))))))...))...((((...((...(((.......)))..))...)))).... ( -36.50) >DroYak_CAF1 26849 111 + 1 UGGUAUUUCAGAACAUAUUCAUGGCCAUGCAGUCAAUGAUCAAGGCCAUGGAUAUGCUGAAGAUUUCCUAUGGUCAGGGAGAGCAUAGUGUAAGUGAAUCCGGAAUCUAAU .......((((..(((((((((((((.((((.....))..)).)))))))))))))))))((((((((((((.((.....)).))))).......(....))))))))... ( -34.50) >consensus UGGUUUUUCAGAACAUAUUCAUGGCCAUGCAGUCAAUGAUCAAGGCCAUGGAUAUGCUGAAGAUUUCCUAUGGUCAGGGAGAGCAUAGUGUAAGUGAAACAUGUAUCUAAU .....((((((..(((((((((((((.((((.....))..)).))))))))))))))))))).((((((......)))))).............................. (-22.63 = -23.47 + 0.84)

| Location | 8,139,904 – 8,140,015 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.10 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
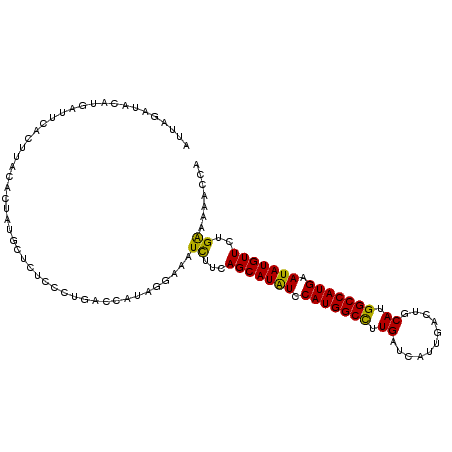
>2R_DroMel_CAF1 8139904 111 - 20766785 AUUAGAUCAAUGUUUCACUUACACUAUGCUCUCCCUGACCGUAGGAAAUCUUCAGCAUAUCCAUGGCCUUGAUCAUUGACUGCAUGGCCAUGAAUAUGUUCUGAAAAACCA .(((((.................(((((.((.....)).)))))..........((((((.(((((((.((...........)).))))))).)))))))))))....... ( -26.70) >DroVir_CAF1 37272 111 - 1 CAUAAUUAUGUUUAGUGCUUACACGAAUCUCUGCACCACCAUAUUCUAUUUUGAGCAUGUCCAUGGCCUUGAUCAUGGACUGCAUGGCCAUGAAUAUGUUUUGGUAGACCA ......((((....((((..............))))...)))).((((((..((((((((.(((((((.((...........)).))))))).)))))))).))))))... ( -30.04) >DroGri_CAF1 18944 111 - 1 CUUAAAUACAUGUAGAGCUUACUUGAAUCUCUGGACCACCAUAUUCUAUUUUGAGCAUAUCCAUGGCUUUGAUCAUGGACUGCAUGGCCAUGAAUAUGUUUUGGAACACCA ........(((((((.(((((...((((...(((....))).)))).....)))))...(((((((......)))))))))))))).(((...........)))....... ( -26.00) >DroSim_CAF1 17097 111 - 1 AUUGGGUAAAUGAUUCACUUACACUAUGCUCUCCCUGACCAUAGGAAAUCUUCAGCAUAUCCAUGGCCUUGAUCAUUGACUGCAUGGCCAUGAAUAUGUUCUGAAAAACCA ....(((................(((((.((.....)).)))))......((((((((((.(((((((.((...........)).))))))).)))))..)))))..))). ( -29.80) >DroEre_CAF1 17112 111 - 1 AUGAGGUCCGUGAUUCACUUACACUAUGUUCUCCCUGACCAUAGGAAAUCUUCAGCAUAUCCAUGGCCUUGAUCAUUGACUGCAUGGCCAUGAAUAUGUUCUGAAAAACCA ..((..(((((((.....))))..((((.((.....)).)))))))..))((((((((((.(((((((.((...........)).))))))).)))))..)))))...... ( -30.70) >DroYak_CAF1 26849 111 - 1 AUUAGAUUCCGGAUUCACUUACACUAUGCUCUCCCUGACCAUAGGAAAUCUUCAGCAUAUCCAUGGCCUUGAUCAUUGACUGCAUGGCCAUGAAUAUGUUCUGAAAUACCA ..........((...........(((((.((.....)).)))))......((((((((((.(((((((.((...........)).))))))).)))))..)))))...)). ( -29.00) >consensus AUUAGAUACAUGAUUCACUUACACUAUGCUCUCCCUGACCAUAGGAAAUCUUCAGCAUAUCCAUGGCCUUGAUCAUUGACUGCAUGGCCAUGAAUAUGUUCUGAAAAACCA ................................................((...(((((((.(((((((.((...........)).))))))).)))))))..))....... (-18.82 = -18.10 + -0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:55 2006