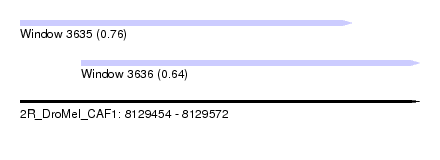
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,129,454 – 8,129,572 |
| Length | 118 |
| Max. P | 0.758602 |

| Location | 8,129,454 – 8,129,552 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -15.87 |
| Consensus MFE | -9.31 |
| Energy contribution | -9.03 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8129454 98 + 20766785 CUC--------------------UCUUCUU-UA-CUUGCAGUUCCACUGCAUACAGAAAAAUAAACAAAUACAAGAAGCAUAAAAUAUGCAGAAAAUUAAAUACAAAAGGAGCCACUAAG ...--------------------.((((((-(.-..(((((.....)))))..........................(((((...)))))...............)))))))........ ( -15.10) >DroVir_CAF1 24077 98 + 1 UUC--------------------UCUAUUU-UU-CUUGCAGUUCCACUGCAUACCGGAAAAUAAACAAAUACAAGAAGCAUAAAGUAUGCAGAAAAUUAAAUACAAAAGAAGCAUUUAAG (((--------------------(.(((((-((-(.(((((.....)))))....))))))))..............(((((...)))))))))..((((((.(.......).)))))). ( -15.80) >DroGri_CAF1 7191 99 + 1 UUC--------------------UCUUUUU-UUCAUUGCAGUUCCACUGCAUACCGGAAAAUAAACAAAUACAAGAAGCAUAAAGUAUGCGGAAAAUUAAAUACAAAAGGAGCAUUUAAG ..(--------------------(((((((-(((..(((((.....))))).....)))..................(((((...)))))..............))))))))........ ( -16.50) >DroWil_CAF1 26687 87 + 1 UUU--------------------UCUUAUC--U-CUUGCAGUUCCAUUGCAUACAGAAAUAUAAACAAAUACAAGAAGUAUAAAGUAUGCAGAAAUUUAAAUACCA----------UAAG ...--------------------.......--.-............(((((((((....)........((((.....))))...))))))))..............----------.... ( -9.70) >DroMoj_CAF1 7171 117 + 1 -UAAUGUAUAUGUAAAUGCAAAUGUUCUUU-UU-AUUGCAGUUCCACUGCAUACCGGAAAAUAAACAAAUACAAGAAGCAUAAAGUAUGCAGAAAAUUAAAUACAAAAGGAGCAUUUAAG -....((((......))))(((((((((((-((-..(((((.....)))))..........................(((((...)))))..............)))))))))))))... ( -23.60) >DroPer_CAF1 8195 99 + 1 CAC--------------------UCUUAUUCCA-CCUGCAGUUCCACUGCAUACAGAAAAAUAAACAAACACAAGAAGCAUAAAGUAUGCAGAAAAUUAAACACAAAAGGAGCCAUUAAG ..(--------------------((((.(((..-..(((((.....)))))....)))...................(((((...))))).................)))))........ ( -14.50) >consensus UUC____________________UCUUUUU_UU_CUUGCAGUUCCACUGCAUACAGAAAAAUAAACAAAUACAAGAAGCAUAAAGUAUGCAGAAAAUUAAAUACAAAAGGAGCAUUUAAG ....................................(((((.....)))))..........................(((((...))))).............................. ( -9.31 = -9.03 + -0.28)

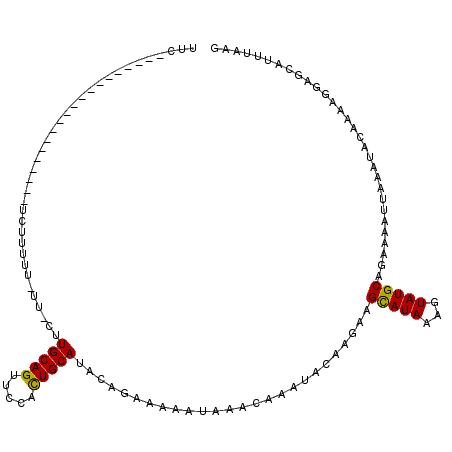

| Location | 8,129,472 – 8,129,572 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.64 |
| Mean single sequence MFE | -16.04 |
| Consensus MFE | -13.42 |
| Energy contribution | -13.23 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8129472 100 + 20766785 GUUCCACUGCAUACAGAAAAAUAAACAAAUACAAGAAGCAUAAAAUAUGCAGAAAAUUAAAUACAAAAGGAGCCACUAAGUGUUUCGUGUGAAAUCAAU-U------------------ (((((.(((....))).....................(((((...)))))..................)))))(((...(.....)..)))........-.------------------ ( -12.90) >DroVir_CAF1 24095 105 + 1 GUUCCACUGCAUACCGGAAAAUAAACAAAUACAAGAAGCAUAAAGUAUGCAGAAAAUUAAAUACAAAAGAAGCAUUUAAGUGUUGCGUGUGAGCGAA-AGUAAAAG------------- ......((((((((..............................)))))))).........((((...(.(((((....))))).).)))).((...-.)).....------------- ( -17.01) >DroPse_CAF1 8113 102 + 1 GUUCCACUGCAUACAGAAAAAUAAACAAACACAAGAAGCAUAAAGUAUGCAGAAAAUUAAACACAAAAGGAGCCAUUAAGUGUUGCGUGUGAAAGAAAU-U-AA--------------- .(((..((((((((..............................)))))))).........((((...(.((((.....).))).).))))...)))..-.-..--------------- ( -16.21) >DroGri_CAF1 7210 119 + 1 GUUCCACUGCAUACCGGAAAAUAAACAAAUACAAGAAGCAUAAAGUAUGCGGAAAAUUAAAUACAAAAGGAGCAUUUAAGUGUUGCGUGUGAAAGUAAAGCAAAAGCGAAGAGAAAAAG .((((.((((((((..............................)))))))).........((((...(.(((((....))))).).))))........((....))...).))).... ( -17.21) >DroMoj_CAF1 7208 106 + 1 GUUCCACUGCAUACCGGAAAAUAAACAAAUACAAGAAGCAUAAAGUAUGCAGAAAAUUAAAUACAAAAGGAGCAUUUAAGUGUUGCGUGUGAAAGAAAAGCAAAAA------------- .(((..((((((((..............................)))))))).........((((...(.(((((....))))).).))))...))).........------------- ( -16.71) >DroPer_CAF1 8214 102 + 1 GUUCCACUGCAUACAGAAAAAUAAACAAACACAAGAAGCAUAAAGUAUGCAGAAAAUUAAACACAAAAGGAGCCAUUAAGUGUUGCGUGUGAAAGAAAU-U-AA--------------- .(((..((((((((..............................)))))))).........((((...(.((((.....).))).).))))...)))..-.-..--------------- ( -16.21) >consensus GUUCCACUGCAUACAGAAAAAUAAACAAAUACAAGAAGCAUAAAGUAUGCAGAAAAUUAAAUACAAAAGGAGCAAUUAAGUGUUGCGUGUGAAAGAAAA_U_AA_______________ ......((((((((..............................)))))))).........((((...(.(((........))).).))))............................ (-13.42 = -13.23 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:47 2006