| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,128,794 – 8,128,907 |
| Length | 113 |
| Max. P | 0.542453 |

| Location | 8,128,794 – 8,128,907 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -27.95 |
| Consensus MFE | -17.81 |
| Energy contribution | -19.77 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
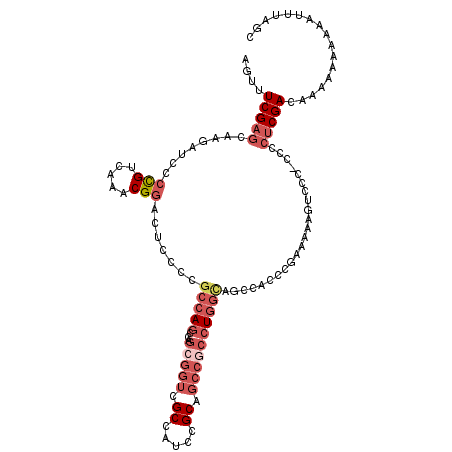
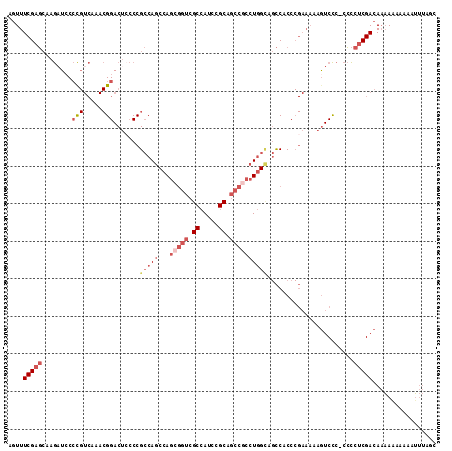
>2R_DroMel_CAF1 8128794 113 + 20766785 AGUUUCGAGCAAGAUCCCCGUCAAACGGACUCCCCGCCAGCCAGCGGUCGCCAUCCGCAGCCGCCUGGCAGCCACCCGAAAAAGUCCC-CCCCUCGACAAAAAAAAAAUUUAGC ..(((((.((.......(((.....))).......(((((...(((((.((.....)).)))))))))).))....)))))..(((..-......)))................ ( -28.60) >DroPse_CAF1 7313 91 + 1 --UUUCGCCAAAAAUCCGU-------GGACACACCGACAGCCAG-----GCAACCAGCAAUCGAAUCG---UCUCC-----UGGUCUC-CGUCUCGACACCACAAUUGGCAGUC --....(((((......(.-------.(((....(((..((..(-----....)..))..)))....)---))..)-----((((.((-......)).))))...))))).... ( -21.60) >DroSec_CAF1 15101 113 + 1 AGUUUCGAGCAAGAUCCCCGUCAAACGGACUCCCCGCCAGCCAGCGGUCGCCAUCCGCAGCCGCCUGGCAGCCACCCGAAAAAGUCCC-CCCCUCGACAAAAAAAAAAUUUAGC ..(((((.((.......(((.....))).......(((((...(((((.((.....)).)))))))))).))....)))))..(((..-......)))................ ( -28.60) >DroSim_CAF1 5956 113 + 1 AGUUUCGAGCAAGAUCCCCGUCAAACGGACUCCCCGCCAGCCAGCGGUCGCCAUCCGCAGCCGCCUGGCAGCCACCCGAAAAAGUCCC-CCCCUCGACAAAAAAAAAAUUUAGC ..(((((.((.......(((.....))).......(((((...(((((.((.....)).)))))))))).))....)))))..(((..-......)))................ ( -28.60) >DroEre_CAF1 5964 113 + 1 AGUUUCGAGCAAGUUACCCGUCAAACGGACUACCCGCCAGCCAGGGGUCGCCAUCCGCGGCCGCCUGGCAGCCACCCGAAAAAGUCCCCCCCCUCGACAAAAA-AAAUUUUAGC ....(((((...(((........)))(((((....((..((((((((((((.....)))))).)))))).))(....)....))))).....)))))......-.......... ( -34.30) >DroYak_CAF1 15451 111 + 1 AGUUUCGAGCAAGAUCCCCGUCAAACGGACUCCCCGCCAGCCAGCGGUCGCCAUCCGCAGCCGCCUGGUAGCCACCCGAAAAAAUCCC-CCCCUCGACAAAA--AAAUUUUAUC ....(((((........(((.....))).......((..(((((((((.((.....)).)))).))))).))................-...))))).....--.......... ( -26.00) >consensus AGUUUCGAGCAAGAUCCCCGUCAAACGGACUCCCCGCCAGCCAGCGGUCGCCAUCCGCAGCCGCCUGGCAGCCACCCGAAAAAGUCCC_CCCCUCGACAAAAAAAAAAUUUAGC ....(((((........(((.....))).......(((((...(((((.((.....)).)))))))))).......................)))))................. (-17.81 = -19.77 + 1.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:46 2006