
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,128,234 – 8,128,392 |
| Length | 158 |
| Max. P | 0.998048 |

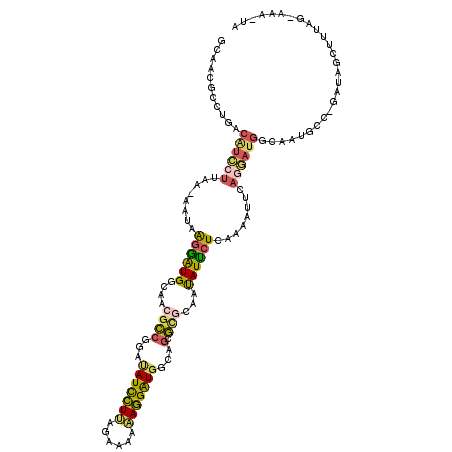
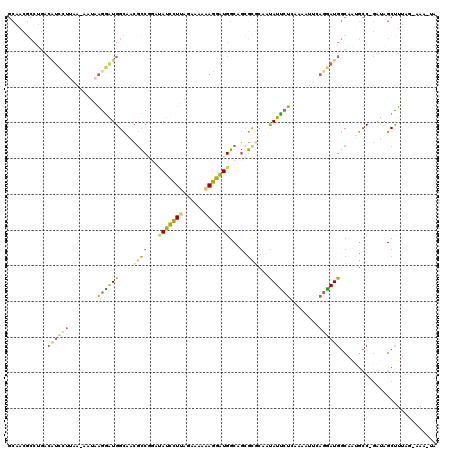
| Location | 8,128,234 – 8,128,352 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.77 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -14.39 |
| Energy contribution | -14.47 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8128234 118 - 20766785 GCAACGCCUUACAUCCUUCA-AAUAAGGAUGGCAACGCCAGACAUCCUUAAAAAUAAGGAUGGCAGUGCGCCAUAUUCUCAUAAUUCAGGAUGGCAAUGCCUGAUAGCUUUAGGAAA-GA (((..(((...(((((((..-...)))))))............(((((((....))))))))))..)))((((..((((........))))))))....(((((.....)))))...-.. ( -34.70) >DroSec_CAF1 14542 117 - 1 GCAACGCCUGACGUCCUUAAAAAUAAGGAUGGCAACGCCGUAUAUCCUUAGAAAAAAGGAUGGCAGCGCGCAGUAUUCUCAAAAUUAAGGAUGGCAAUGCC-GAUAGCCUUAG-AAU-UA ((..((((((.(((((((.....((((((((((...)))....))))))).....))))))).))).)))..))..........((((((.((((...)))-)....))))))-...-.. ( -35.70) >DroSim_CAF1 5402 116 - 1 GCAACGCCUGACGUCCUUAAAAAUAAGGAUGGCAACGCCGUAUAUCCUUAGAAA-AAGGAUGGCAGCGCGCAGUAUUCUCUAAAUUUAGGAUGGCAAUGCC-GAUAGCCUUAG-AAU-UA ((..((((((.(((((((.....((((((((((...)))....)))))))....-))))))).))).)))..))....(((((..(((...((((...)))-).)))..))))-)..-.. ( -33.10) >DroEre_CAF1 5471 92 - 1 ------------------------UGGGGUGGCAGCACCGGAUAUUAUAAACAUAUAUGGUGGCAGCGCCCGAUAUCCU---AAAUCCGGAUGACAGCGACCUACAGCUUUAA-AAAAUA ------------------------..((((.((..((((((((.(((.....((((..((((....))))..))))..)---)))))))).))...)).))))..........-...... ( -23.10) >DroYak_CAF1 14897 116 - 1 GCAACACCCGGCAUUCUUAACAAAUAAAGUGGUAGCGUCCGAAAUCCUUAGAAAUCAGGAUGGCAUUAUAAAACAUUCUCAAAAAUCAUAAUGGCAACACCCGACAGCUUUAA-AAA--- ((........))............((((((.((.......(..(((((.(....).)))))..)...........................((....))....)).)))))).-...--- ( -15.00) >consensus GCAACGCCUGACAUCCUUAA_AAUAAGGAUGGCAACGCCGGAUAUCCUUAGAAAAAAGGAUGGCAGCGCGCAAUAUUCUCAAAAUUCAGGAUGGCAAUGCC_GAUAGCUUUAG_AAA_UA ...........((((((........((((((....((((...(((((((......)))))))...).)))...))))))........))))))........................... (-14.39 = -14.47 + 0.08)

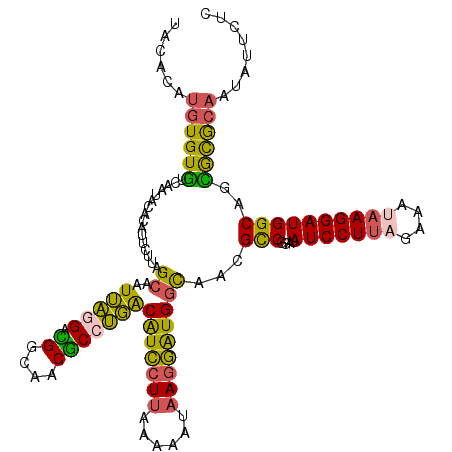
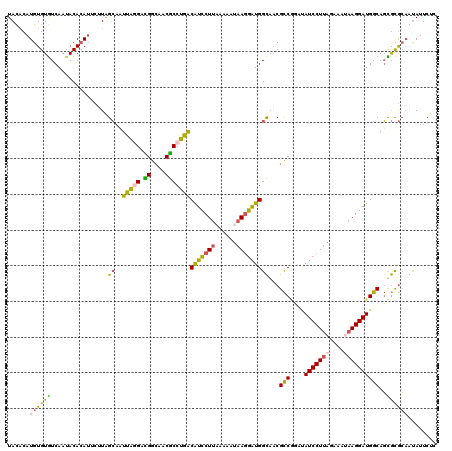
| Location | 8,128,273 – 8,128,392 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -26.31 |
| Energy contribution | -25.25 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8128273 119 - 20766785 UAUUCAUGUGUGUCAAUACACAUUCUUAGCAAUAAGGUUGGCAACGCCUUACAUCCUUCA-AAUAAGGAUGGCAACGCCAGACAUCCUUAAAAAUAAGGAUGGCAGUGCGCCAUAUUCUC .....(((((((....)))))))(((((....))))).((((.........(((((((..-...)))))))(((..((((....((((((....))))))))))..)))))))....... ( -38.00) >DroSec_CAF1 14579 120 - 1 UACACAUGUGUGUCAAUACACAUUCUUAGCAUUUAGGACGGCAACGCCUGACGUCCUUAAAAAUAAGGAUGGCAACGCCGUAUAUCCUUAGAAAAAAGGAUGGCAGCGCGCAGUAUUCUC (((..(((((((....))))))).....((..(((((.((....)))))))((((((((....))))))))))...(((((.(((((((......)))))))...))).)).)))..... ( -39.40) >DroSim_CAF1 5439 119 - 1 UACACAUGUGUGUCAAAACACAUUCUUGGCAUUUACGACGGCAACGCCUGACGUCCUUAAAAAUAAGGAUGGCAACGCCGUAUAUCCUUAGAAA-AAGGAUGGCAGCGCGCAGUAUUCUC (((.((.((((((....))))))...))((.........(....)(((((.(((((((.....((((((((((...)))....)))))))....-))))))).))).)))).)))..... ( -35.50) >DroYak_CAF1 14933 118 - 1 UACACAUGUGUAUCAAUACAAA--CUUAGGAAUUGUGAUGGCAACACCCGGCAUUCUUAACAAAUAAAGUGGUAGCGUCCGAAAUCCUUAGAAAUCAGGAUGGCAUUAUAAAACAUUCUC ......(((((....)))))..--....((((((((((((.(......(((.((.((..((.......))...)).)))))..(((((.(....).)))))).)))))))....))))). ( -20.30) >consensus UACACAUGUGUGUCAAUACACAUUCUUAGCAAUUAGGACGGCAACGCCUGACAUCCUUAAAAAUAAGGAUGGCAACGCCGGAUAUCCUUAGAAAUAAGGAUGGCAGCGCGCAAUAUUCUC ......((((((................((..(((((.((....)))))))(((((((......)))))))))...(((....(((((((....))))))))))..))))))........ (-26.31 = -25.25 + -1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:45 2006