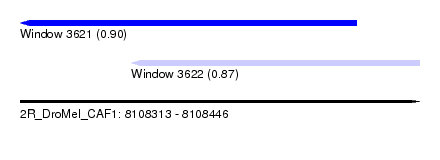
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,108,313 – 8,108,446 |
| Length | 133 |
| Max. P | 0.902446 |

| Location | 8,108,313 – 8,108,425 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.47 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -11.66 |
| Energy contribution | -12.58 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
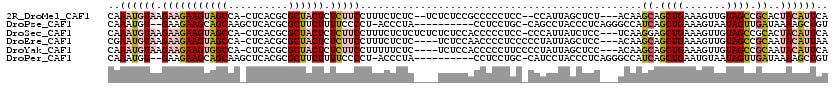
>2R_DroMel_CAF1 8108313 112 - 20766785 CAAAUGUAAGAAGAAGUAGCCA-CUCACGCGCUACUCUCUUCCUUUCUCUC--UCUCUCCGCCCCCUCC--CCAUUAGCUCU---ACAAGCAGCUGAAAGUUGUAGCCGCACUACAUUCA ..((((((.((((((((((((.-.....).)))))).))))).........--................--......((.((---((((.(........)))))))..))..)))))).. ( -21.90) >DroPse_CAF1 24460 106 - 1 CAAAUGU--GAAGAAGCAGCAAGCUCACGCGCUUCUUUUCCCCU-ACCCUA----------CCUCCUGC-CAGCCUACCCUCAGGGCCAUCAGCUGAAAGUAAUAGUUGAUAAAAGCUGU .......--((((((((.((........))))))))))......-......----------........-((((...(((...)))..((((((((.......))))))))....)))). ( -23.70) >DroSec_CAF1 23342 115 - 1 CAAAUGUAAGAAGAAGUAGCCA-CUCACGCGCUACUCUCUUCCUUUCUCUCUCUCUCUCCACCCCCUCC-CCCAUUAUCUCC---UCAAGGAGCUGAAAGUUGUAGCCGCACUACAUUCA .........((((((((((((.-.....).)))))).)))))...........................-....(((.((((---....)))).)))....(((((.....))))).... ( -21.80) >DroEre_CAF1 23555 112 - 1 CGAAUGUAAGAAGAAGUAGCCA-CUCACGCGCUACUCUCUUCCUUUCUCUC----UCUCCAACCCCUCCCCCUAUUAGCUCC---ACAAGCAGCUGAAAGUUGUAGCCGCAAUACAUUAA ..((((((.((((((((((((.-.....).)))))).))))).........----................((.((((((.(---....).)))))).))((((....)))))))))).. ( -22.90) >DroYak_CAF1 23484 112 - 1 CAAAUGUAAGAAGAAGUGGCCA-CUCACGCGCUACUCUCUUCCUUUUUCUC----UCUCCACCCCCUUCCCCUAUUAGCUCC---ACAAGCAGCUGAAAGUUGUAGCCGCAAUACAUUCA ..((((((.((((((((((((.-.....).)))))).))))).........----................((.((((((.(---....).)))))).))((((....)))))))))).. ( -21.90) >DroPer_CAF1 24473 106 - 1 CAAAUGU--GAAGAAGCAGCAAGCUCACGCGCUUCUUUUCCCCU-ACCCUA----------CCUCCUGC-CAUCCUACCCUCAGGGCCAUCAGCUGAAUGUAAUAGUUGAUAAAAGCUGU .......--((((((((.((........))))))))))......-......----------......((-..((((......))))..((((((((.......))))))))....))... ( -20.10) >consensus CAAAUGUAAGAAGAAGUAGCCA_CUCACGCGCUACUCUCUUCCUUUCUCUC____UCUCCACCCCCUCC_CCUAUUAGCUCC___ACAAGCAGCUGAAAGUUGUAGCCGCAAUACAUUCA ..((((((.(((((((((((..........)))))).)))))...............................................((.((((.......)))).))..)))))).. (-11.66 = -12.58 + 0.92)

| Location | 8,108,350 – 8,108,446 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -19.02 |
| Consensus MFE | -14.47 |
| Energy contribution | -14.67 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
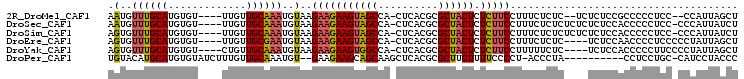
>2R_DroMel_CAF1 8108350 96 - 20766785 AAUGUUUGCAUGUGU----UUGUUGCAAAUGUAAGAAGAAGUAGCCA-CUCACGCGCUACUCUCUUCCUUUCUCUC--UCUCUCCGCCCCCUCC--CCAUUAGCU .(((((((((.....----....)))))))))..((((((((((((.-.....).)))))).))))).........--................--......... ( -19.70) >DroSec_CAF1 23379 99 - 1 AAUGUUUGCAUGUGU----UUGUUGCAAAUGUAAGAAGAAGUAGCCA-CUCACGCGCUACUCUCUUCCUUUCUCUCUCUCUCUCCACCCCCUCC-CCCAUUAUCU .(((((((((.....----....)))))))))..((((((((((((.-.....).)))))).)))))...........................-.......... ( -19.70) >DroSim_CAF1 26297 99 - 1 AGUGUUUGCAUGUGU----UUGUUGCAAAUGUAAGAAGAAGUAGCCA-CUCACGCGCUACUCUCUUCCUUUCUCUCUCUCUCUCCACCCCCUCC-CCCAUUAUCU .(((.((((((.(((----.....))).))))))((((((((((((.-.....).)))))).))))).................))).......-.......... ( -20.40) >DroEre_CAF1 23592 96 - 1 AGUGUUUGCAUGUGU----UUGUUGCGAAUGUAAGAAGAAGUAGCCA-CUCACGCGCUACUCUCUUCCUUUCUCUC----UCUCCAACCCCUCCCCCUAUUAGCU .(..((((((.....----....))))))..)..((((((((((((.-.....).)))))).))))).........----......................... ( -19.00) >DroYak_CAF1 23521 96 - 1 AGUGUUUGCAUGUGU----CUGUUGCAAAUGUAAGAAGAAGUGGCCA-CUCACGCGCUACUCUCUUCCUUUUUCUC----UCUCCACCCCCUUCCCCUAUUAGCU .(((.((((((.(((----.....))).))))))((((((((((((.-.....).)))))).))))).........----....))).................. ( -19.30) >DroPer_CAF1 24513 91 - 1 UGUACAUGCAUGUGUAUCUUUGUUGCAAAUGU--GAAGAAGCAGCAAGCUCACGCGCUUCUUUUCCCCU-ACCCUA----------CCUCCUGC-CAUCCUACCC .......((((.((((.......)))).))))--((((((((.((........))))))))))......-......----------........-.......... ( -16.00) >consensus AGUGUUUGCAUGUGU____UUGUUGCAAAUGUAAGAAGAAGUAGCCA_CUCACGCGCUACUCUCUUCCUUUCUCUC____UCUCCACCCCCUCC_CCCAUUAGCU .(..((((((.............))))))..)..(((((((((((..........)))))).)))))...................................... (-14.47 = -14.67 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:34 2006