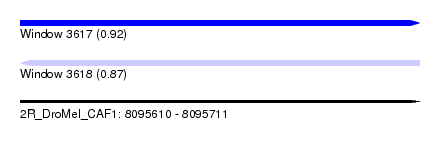
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,095,610 – 8,095,711 |
| Length | 101 |
| Max. P | 0.916083 |

| Location | 8,095,610 – 8,095,711 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -14.52 |
| Energy contribution | -15.10 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
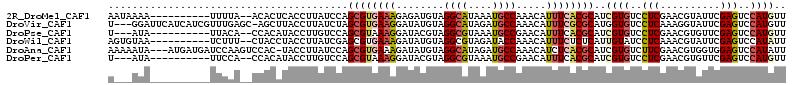
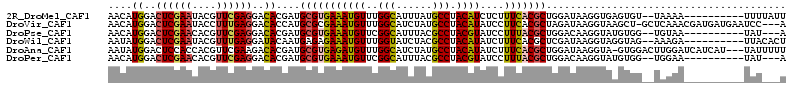
>2R_DroMel_CAF1 8095610 101 + 20766785 AAUAAAA----------UUUUA--ACACUCACCUUAUCCAGCGUGAAAGAGAUGUAGGCAUAAAUGCCAAACAUUUCACGCAUCGUGUCCUCGAACGUAUUCGAGUCCAUGUU .......----------.....--................((((((....(((((.((((....))))..)))))))))))..((((..((((((....))))))..)))).. ( -28.10) >DroVir_CAF1 12933 109 + 1 U---GGAUUCAUCAUCGUUUGAGC-AGCUUACCUUAUCUAGCGUGAAGGAUAUGUAGGCAUAGAUGCCAAACAUUUCGCGCAUGGUGUCCUCAAAGGUAUUCGAGUCCAUGUU (---((((((.....(.((((((.-.((((((..(((((........))))).))))))...(((((((..(.......)..))))))))))))).).....))))))).... ( -33.30) >DroPse_CAF1 10638 98 + 1 U---AUA----------UUACA--CCACAUACCUUGUCCAGCGUAAAGGAUACGUAGGCGUAAAUGCCGAACAUUUCACGCAUCGUGUCCUCGAACGUGUUCGAGUCCAUGUU .---...----------.....--..((((..((((..((.(((..((((((((...(((((((((.....))))).))))..))))))))...)))))..))))...)))). ( -29.50) >DroWil_CAF1 12449 101 + 1 AGUGUAA----------UCUUU--CUACCUACCUUAUCGAGCGUGAAAGAUAUGUAGGCGUAGAUACCAAACAUUUCUCUCAUUGUAUCCUCAAACGUAUUCGAGUCCAUAUU (((((..----------....(--((((((((..((((..........)))).)))))..))))......)))))...(((...((((........))))..)))........ ( -13.80) >DroAna_CAF1 10427 109 + 1 AAAAAUA---AUGAUGAUCCAAGUCCAC-UACCUUAUCCAGCGUGAAAGAUAUGUAGGCAUAGAUGCCAAACAUCUCACGCAUCGUGUCUUCGAACGUGGUGGAGUCCAUAUU .......---.............(((((-(((........((((((.....((((.((((....))))..)))).)))))).(((......)))..))))))))......... ( -28.50) >DroPer_CAF1 10602 98 + 1 U---AUA----------UUCCA--CCACAUACCUUGUCCAGCGUAAAGGAUACGUAGGCGUAAAUGCCGAACAUUUCACGCAUCGUGUCCUCGAACGUGUUCGAGUCCAUGUU .---...----------.....--..((((..((((..((.(((..((((((((...(((((((((.....))))).))))..))))))))...)))))..))))...)))). ( -29.50) >consensus A___AUA__________UUUCA__CCACAUACCUUAUCCAGCGUGAAAGAUAUGUAGGCAUAAAUGCCAAACAUUUCACGCAUCGUGUCCUCGAACGUAUUCGAGUCCAUGUU ........................................((((((((........((((....)))).....))))))))..((((..(((..........)))..)))).. (-14.52 = -15.10 + 0.58)
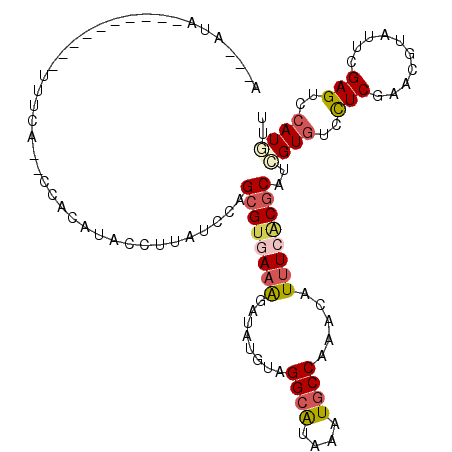
| Location | 8,095,610 – 8,095,711 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -19.46 |
| Energy contribution | -19.43 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
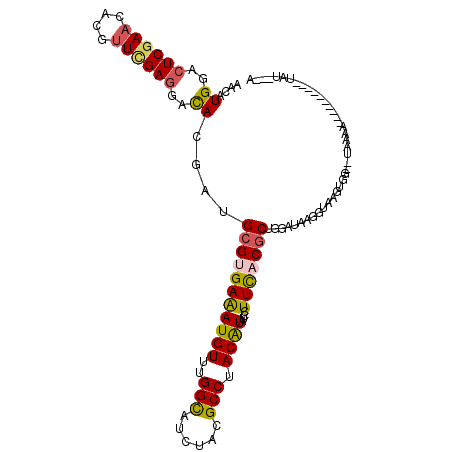
>2R_DroMel_CAF1 8095610 101 - 20766785 AACAUGGACUCGAAUACGUUCGAGGACACGAUGCGUGAAAUGUUUGGCAUUUAUGCCUACAUCUCUUUCACGCUGGAUAAGGUGAGUGU--UAAAA----------UUUUAUU ..(.((..((((((....))))))..)).)..(((((((((((..((((....)))).))))....)))))))..((((((((......--....)----------))))))) ( -29.20) >DroVir_CAF1 12933 109 - 1 AACAUGGACUCGAAUACCUUUGAGGACACCAUGCGCGAAAUGUUUGGCAUCUAUGCCUACAUAUCCUUCACGCUAGAUAAGGUAAGCU-GCUCAAACGAUGAUGAAUCC---A ..(((((.((((((....))))))....)))))........((((((((.((.(((((...((((..........))))))))))).)-)).)))))(((.....))).---. ( -25.40) >DroPse_CAF1 10638 98 - 1 AACAUGGACUCGAACACGUUCGAGGACACGAUGCGUGAAAUGUUCGGCAUUUACGCCUACGUAUCCUUUACGCUGGACAAGGUAUGUGG--UGUAA----------UAU---A ....((..((((((....))))))..)).(((((.(((.....))))))))((((((.(((((.((((..........)))))))))))--)))).----------...---. ( -29.80) >DroWil_CAF1 12449 101 - 1 AAUAUGGACUCGAAUACGUUUGAGGAUACAAUGAGAGAAAUGUUUGGUAUCUACGCCUACAUAUCUUUCACGCUCGAUAAGGUAGGUAG--AAAGA----------UUACACU ..(((.((((((((....)))))).......(((((((.((((..(((......))).)))).)))))))...)).)))..((((....--.....----------))))... ( -20.70) >DroAna_CAF1 10427 109 - 1 AAUAUGGACUCCACCACGUUCGAAGACACGAUGCGUGAGAUGUUUGGCAUCUAUGCCUACAUAUCUUUCACGCUGGAUAAGGUA-GUGGACUUGGAUCAUCAU---UAUUUUU ......((.((((((((..(((......))).(((((((((((..((((....))))...)))))..))))))...........-))))...)))))).....---....... ( -28.70) >DroPer_CAF1 10602 98 - 1 AACAUGGACUCGAACACGUUCGAGGACACGAUGCGUGAAAUGUUCGGCAUUUACGCCUACGUAUCCUUUACGCUGGACAAGGUAUGUGG--UGGAA----------UAU---A ..(.((..((((((....))))))..)).)..(((((((.((.....)))))))))(((((((.((((..........)))))))))))--.....----------...---. ( -27.90) >consensus AACAUGGACUCGAACACGUUCGAGGACACGAUGCGUGAAAUGUUUGGCAUCUACGCCUACAUAUCCUUCACGCUGGAUAAGGUAAGUGG__UAAAA__________UAU___A ....((..((((((....))))))..))....(((((((((((..(((......))).))))....)))))))........................................ (-19.46 = -19.43 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:31 2006