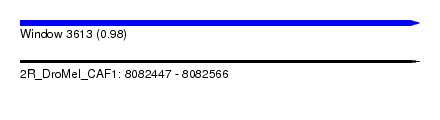
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,082,447 – 8,082,566 |
| Length | 119 |
| Max. P | 0.984877 |

| Location | 8,082,447 – 8,082,566 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -18.23 |
| Energy contribution | -19.23 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8082447 119 + 20766785 UCCACUUCCGAGUGGCAAAACGCUCUGCCCUACAAUCAAAUACCCGGACCUAAGCCCAUCCCCAUUCUGGGCAAUACAUGGAGGUAACCGAAAACCAAACCAAACCAAAACCCAUCAUC ...(((((((((((......)))))............................(((((.........))))).......)))))).................................. ( -23.00) >DroSim_CAF1 184 114 + 1 UCCACUUCCGAGUGGCAAAACGCUCUGCCCUACAAUCAGAUUCCCGGACCUAAGCCCAUCCCCAUUCUGGGCAAUACAUGGAGGUAACCGAAAACCAAACCA-----AAACCGAUCAUC ...(((((((((((......)))))(((((........(((....((........)))))........)))))......)))))).................-----............ ( -23.89) >DroEre_CAF1 184 108 + 1 UCCACUUCCGAGUGGCAAAACGCUCUGCCCUACAAUCAAAUUCCCGGACCCAAGCCCAUUCCUAUUUUGGGCAAUACAUGGAGGUAAU--AAAACC---------GAUAACCCAUCAAC ...(((((((((((......)))))............................(((((.........))))).......))))))...--......---------(((.....)))... ( -24.00) >DroYak_CAF1 184 107 + 1 UCCACUUCCGAGUGGCAAAACGCUCUGCCCUACAAUCAGAUUCCCGGCCCUAAGCCCAUUCCUAUUCUGGGCAAUACCUGGAGGUAAU--AAAA-C---------GAAAACCCAUCAUC ...(((((((((((......)))))(((((...(((..((.....(((.....)))...))..)))..)))))......))))))...--....-.---------.............. ( -26.90) >DroAna_CAF1 214 96 + 1 GCCACAUCAGAGUGGCAGAAUGCCCUGCCCUACAAUCAGAUACCAGGACCAAAGCCCAUACCCAUUUUGGGCAACACCUGGCGGUUAG--UAU---------------------UCGAC (((((......))))).((((((...(((((......))...(((((......(((((.........)))))....))))).)))..)--)))---------------------))... ( -32.00) >DroPer_CAF1 199 111 + 1 UCCACUCCCGAAUGGCAGAACGCUCUGCCCUACAAUCAGAUACCUGGACCCAAGCCCCUACCCAUUCUGGGCAACACCUGGCGGUAGG--UACU-CGUAUAA-----GAGUCGCGACAC (((((((......((((((....)))))).(((..((((....)))).....((..((((((....(..((.....))..).))))))--..))-.)))...-----)))).).))... ( -29.50) >consensus UCCACUUCCGAGUGGCAAAACGCUCUGCCCUACAAUCAGAUACCCGGACCUAAGCCCAUACCCAUUCUGGGCAAUACAUGGAGGUAAC__AAAA_C___________AAACCCAUCAAC ...(((((((.((((((........))))........................(((((.........)))))...)).))))))).................................. (-18.23 = -19.23 + 1.00)

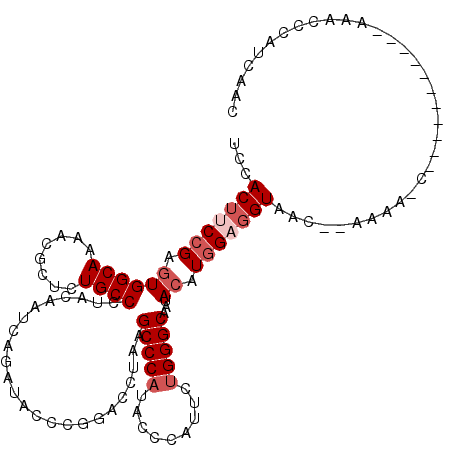

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:26 2006