
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,066,102 – 8,066,211 |
| Length | 109 |
| Max. P | 0.849272 |

| Location | 8,066,102 – 8,066,199 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.29 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.75 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
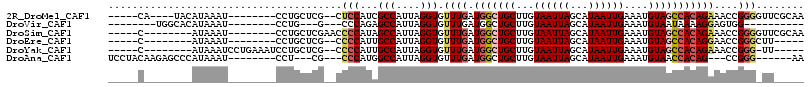
>2R_DroMel_CAF1 8066102 97 + 20766785 -----CA----UACAUAAAU--------CCUGCUCG--CUCCAUCGCCAUUAGGUGUUUGAUGGCUGCUUGUAAUUAGCAUAAUUGAAAUGUAGCCACAGAAACCGGGGUUCGCAA -----..----.........--------..(((..(--((((..((((....))))((((.(((((((...((((((...))))))....)))))))))))....)))))..))). ( -29.10) >DroVir_CAF1 16620 84 + 1 --------UGGCACAUAAAU--------CCUG---G---CCCAGAGCCAUUAGGUGUUUGAUGGCUGCUUGUAAUUAGCAUAAUUGAAAUGUAAUAAAAGGAGUGG---------- --------...(((.....(--------(((.---.---.....(((((((((....)))))))))(((.......)))...................))))))).---------- ( -17.00) >DroSim_CAF1 15154 95 + 1 -----C--------AUAAAU--------CCUGCUCGAACCCCAUAGCCAUUAGGUGUUUGAUGGCUGCUUGUAAUUAGCAUAAUUGAAAUGUAGCCACAGAAACCGGGGUUCGCAA -----.--------......--------..(((..(((((((...(((....))).((((.(((((((...((((((...))))))....)))))))))))....)))))))))). ( -32.80) >DroEre_CAF1 29548 88 + 1 -----C--------AUAAAU--------CCUGCUCG--CCCCAUUGCCAUUAGGUGUUUGAUGGCUGCUUGUAAUUAGCAUAAUUGAAAUGUAGCCACAGGAACCGGGCUU----- -----.--------......--------...(((((--(......)).....(((.((((.(((((((...((((((...))))))....))))))))))).)))))))..----- ( -24.60) >DroYak_CAF1 29477 95 + 1 -----C--------AUAAAUCCUGAAAUCCUGCUCG--CCCCAUUGCCAUUAGGUGUUUGAUGGCUGCUUGUAAUUAGCAUAAUUGAAAUGUAGCCACAGAAACCGGG-UU----- -----.--------...........(((((.....(--(......)).....(((.((((.(((((((...((((((...))))))....))))))))))).))))))-))----- ( -23.60) >DroAna_CAF1 30154 93 + 1 UCCUACAAGAGCCCAUAAAU--------CCU---CG---CCCAUGGCCAUUAGGUGUUUGAUGGCUGCUUGUAAUUAGCAUAAUUGAAAUGUAACCACAG---CCGGG------AA (((((((((((((..(((((--------(((---.(---((...)))....))).)))))..)))).))))).....((((.......))))........---..)))------). ( -21.50) >consensus _____C________AUAAAU________CCUGCUCG__CCCCAUAGCCAUUAGGUGUUUGAUGGCUGCUUGUAAUUAGCAUAAUUGAAAUGUAGCCACAGAAACCGGG_UU_____ .......................................(((...(((....))).((((.(((((((...((((((...))))))....)))))))))))....)))........ (-15.00 = -15.75 + 0.75)
| Location | 8,066,121 – 8,066,211 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 83.70 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -22.36 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
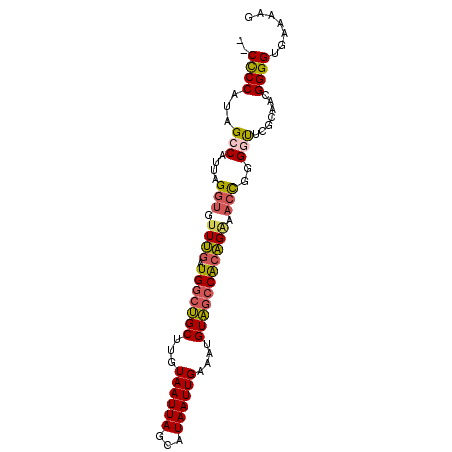
>2R_DroMel_CAF1 8066121 90 + 20766785 --CUCCAUCGCCAUUAGGUGUUUGAUGGCUGCUUGUAAUUAGCAUAAUUGAAAUGUAGCCACAGAAACCGGGGUUCGCAACGGGGUGAAAAG --.....(((((........((((.(((((((...((((((...))))))....)))))))))))..(((..(....)..)))))))).... ( -27.70) >DroSec_CAF1 31240 92 + 1 AACCCCAUAGCCAUUAGGUGUUUGAUGGCUGCUUGUAAUUAGCAUAAUUGAAAUGUAGCCACAGAAACCGGGGUUCGCAACGGGGUGAAAAG .(((((..((((....(((.((((.(((((((...((((((...))))))....))))))))))).)))..))))......)))))...... ( -31.20) >DroSim_CAF1 15169 92 + 1 AACCCCAUAGCCAUUAGGUGUUUGAUGGCUGCUUGUAAUUAGCAUAAUUGAAAUGUAGCCACAGAAACCGGGGUUCGCAACGGGGUGAAAAG .(((((..((((....(((.((((.(((((((...((((((...))))))....))))))))))).)))..))))......)))))...... ( -31.20) >DroEre_CAF1 29563 84 + 1 --CCCCAUUGCCAUUAGGUGUUUGAUGGCUGCUUGUAAUUAGCAUAAUUGAAAUGUAGCCACAGGAACCGGGCUU------GGGGUGAAAAG --(((((..(((....(((.((((.(((((((...((((((...))))))....))))))))))).))).))).)------))))....... ( -33.40) >DroYak_CAF1 29500 83 + 1 --CCCCAUUGCCAUUAGGUGUUUGAUGGCUGCUUGUAAUUAGCAUAAUUGAAAUGUAGCCACAGAAACCGGG-UU------GGGGUGGAAAG --(((((..(((....(((.((((.(((((((...((((((...))))))....))))))))))).))).))-))------))))....... ( -31.50) >DroPer_CAF1 4635 82 + 1 ---CCCAUGGCCAUUAGGUGUUCGAUGGCCGCUUGUAAUUAGCAUAAUUGAAAUGUGUACGGGG----UGGGGCAUGCGGCGGG---GGCAC ---((((((((((((........))))))).((((((....((((.......)))).)))))))----))))...(((......---.))). ( -26.60) >consensus __CCCCAUAGCCAUUAGGUGUUUGAUGGCUGCUUGUAAUUAGCAUAAUUGAAAUGUAGCCACAGAAACCGGGGUUCGCAACGGGGUGAAAAG ..((((...(((....(((.((((.(((((((...((((((...))))))....))))))))))).)))..))).......))))....... (-22.36 = -22.52 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:16 2006