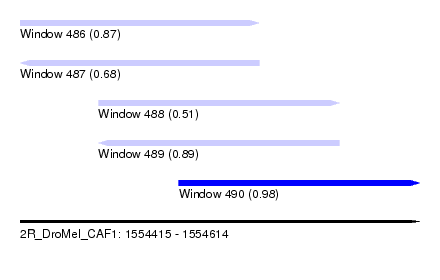
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,554,415 – 1,554,614 |
| Length | 199 |
| Max. P | 0.978015 |

| Location | 1,554,415 – 1,554,534 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.69 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -28.64 |
| Energy contribution | -29.52 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1554415 119 + 20766785 UUGUGCUCAAUUGUGCCAUGAAGGCCCAUAAGGUUUCAC-UGCCCAUGGGCUUGUAAGCUGUGCAAAUGUUUAUUUCUGCGAUUCAGGUCAAAACAUUUGACCAGGUUGCAUCUGGUCAA ..........(((.((((.((((((((((..(((.....-.))).))))))))((((.(((..(((((((((...((((.....))))...)))))))))..))).)))).))))))))) ( -40.00) >DroSec_CAF1 4238 119 + 1 UUGUGCUCAAUUGUGCAUUGAAGGCCCAUAAGGUUCCAC-UGCCCAUGGGCUUGUUAGCUGUGCAAAUGUUUUUUUCUGCGAUUCAGGUCAAAACACUUGACCAGGUCGCAUCUGGUCAA ..........(((..((((((((((((((..(((.....-.))).)))))))).)))).))..))).((((((..((((.....))))..))))))..(((((((((...))))))))). ( -39.80) >DroSim_CAF1 4724 118 + 1 UUGUGCUCAAUUGUGCAUUGAAGGCCCACAAGGUUCCAC-UGCCCAUGGGCUUGUUAGCUGUGCAAAUGUUUUUUUCUGCGAUUCAGGUCA-AACACUUGACCAGGUCGCAUCUGGUCAA ...((..((.(((..(((((((((((((...(((.....-.)))..))))))).)))).))..)))...........(((((((..(((((-(....)))))).)))))))..))..)). ( -37.50) >DroEre_CAF1 4657 118 + 1 CUGGGCUCAAUUGUGCAUUGGCGGCC-UAAAGGUUCCAC-UGCCCCUGGGCUCGUAAGCUGUGCAAAUGUUUAUUUCUGCGAUUCAGGUCAAGCCAUUUGACCAGGUUGCAUCUGGUCAA ...((((((.(((..((...((((((-((..(((.....-.)))..))))).)))....))..))).))......((((.....))))...))))..((((((((((...)))))))))) ( -39.90) >DroYak_CAF1 10224 120 + 1 UUGGGCUCACUUGCGCAUUAAAGGCCUUAAAGGUUUCAUUUGGCAAUGGGCUUCUAAGCUGUGCAAAUGUUUAUUUCUGCGAUUCAGGUCAAGACAUUUGACCAGGUUGCAUCUGAUCAA ..(((((((.((((.((....(((((.....)))))....))))))))))))).(((.(((..(((((((((...((((.....))))...)))))))))..))).)))........... ( -33.20) >consensus UUGUGCUCAAUUGUGCAUUGAAGGCCCAAAAGGUUCCAC_UGCCCAUGGGCUUGUAAGCUGUGCAAAUGUUUAUUUCUGCGAUUCAGGUCAAAACAUUUGACCAGGUUGCAUCUGGUCAA .......((.(((((((....(((((((...(((.......)))..)))))))......))))))).))........(((((((..((((((.....)))))).)))))))......... (-28.64 = -29.52 + 0.88)

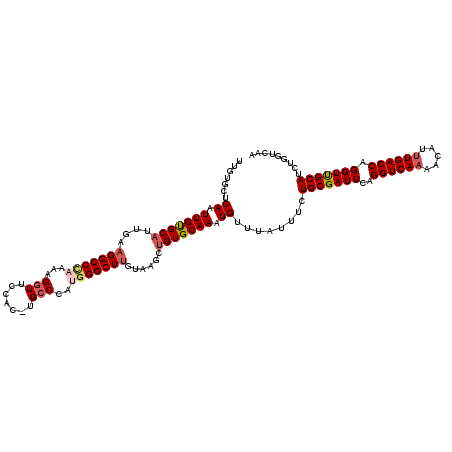
| Location | 1,554,415 – 1,554,534 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.69 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.26 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1554415 119 - 20766785 UUGACCAGAUGCAACCUGGUCAAAUGUUUUGACCUGAAUCGCAGAAAUAAACAUUUGCACAGCUUACAAGCCCAUGGGCA-GUGAAACCUUAUGGGCCUUCAUGGCACAAUUGAGCACAA ..((((((.......))))))((((((((....(((.....)))....)))))))).....(((((...(((....))).-((....((....))(((.....)))))...))))).... ( -34.00) >DroSec_CAF1 4238 119 - 1 UUGACCAGAUGCGACCUGGUCAAGUGUUUUGACCUGAAUCGCAGAAAAAAACAUUUGCACAGCUAACAAGCCCAUGGGCA-GUGGAACCUUAUGGGCCUUCAAUGCACAAUUGAGCACAA ((((((((.......))))))))(((((((...(((.....)))...))))))).(((...........(((((((((..-......)).)))))))..(((((.....))))))))... ( -35.70) >DroSim_CAF1 4724 118 - 1 UUGACCAGAUGCGACCUGGUCAAGUGUU-UGACCUGAAUCGCAGAAAAAAACAUUUGCACAGCUAACAAGCCCAUGGGCA-GUGGAACCUUGUGGGCCUUCAAUGCACAAUUGAGCACAA ((((((((.......))))))))(((((-..(..((....(((((........)))))...((......(((((..((..-.......))..))))).......)).)).)..))))).. ( -36.92) >DroEre_CAF1 4657 118 - 1 UUGACCAGAUGCAACCUGGUCAAAUGGCUUGACCUGAAUCGCAGAAAUAAACAUUUGCACAGCUUACGAGCCCAGGGGCA-GUGGAACCUUUA-GGCCGCCAAUGCACAAUUGAGCCCAG ((((((((.......))))))))..((((..(((((....(((((........)))))...(((....))).)))).(((-.(((..((....-))...))).)))....)..))))... ( -39.90) >DroYak_CAF1 10224 120 - 1 UUGAUCAGAUGCAACCUGGUCAAAUGUCUUGACCUGAAUCGCAGAAAUAAACAUUUGCACAGCUUAGAAGCCCAUUGCCAAAUGAAACCUUUAAGGCCUUUAAUGCGCAAGUGAGCCCAA ((((((((.......))))))))..........(((.....))).......(((((((.((..(((((.(((((((....))))..........))).))))))).)))))))....... ( -25.20) >consensus UUGACCAGAUGCAACCUGGUCAAAUGUUUUGACCUGAAUCGCAGAAAUAAACAUUUGCACAGCUUACAAGCCCAUGGGCA_GUGGAACCUUAUGGGCCUUCAAUGCACAAUUGAGCACAA ((((((((.......)))))))).(((.............(((((........)))))...(((((...(((....)))...((((.((.....))..)))).........)))))))). (-24.22 = -24.26 + 0.04)

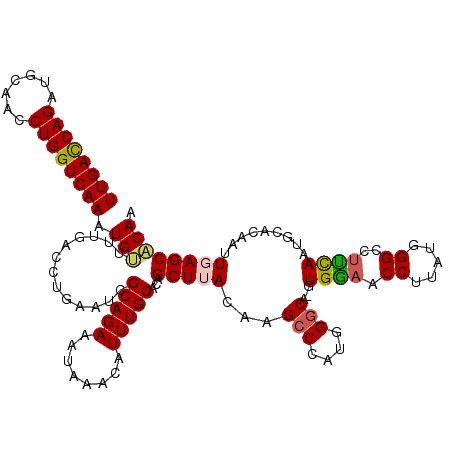
| Location | 1,554,454 – 1,554,574 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -34.16 |
| Consensus MFE | -25.42 |
| Energy contribution | -25.70 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1554454 120 + 20766785 UGCCCAUGGGCUUGUAAGCUGUGCAAAUGUUUAUUUCUGCGAUUCAGGUCAAAACAUUUGACCAGGUUGCAUCUGGUCAAAUAGAUCUCGAGCAAAGCGGGCACCUAUAAAUUAACGAUC (((((.(..(((((....((((..((((((((...((((.....))))...))))))))((((((((...))))))))..))))....)))))..)..)))))................. ( -36.70) >DroSec_CAF1 4277 120 + 1 UGCCCAUGGGCUUGUUAGCUGUGCAAAUGUUUUUUUCUGCGAUUCAGGUCAAAACACUUGACCAGGUCGCAUCUGGUCAAAUAGAUCUCGAGCAAAGCGAGCACAUAUAAAUUAGCGAUC .(((....)))(((((((.(((((...((((((.((((((((((..((((((.....)))))).)))))))...((((.....))))..))).)))))).)))))......))))))).. ( -35.60) >DroSim_CAF1 4763 119 + 1 UGCCCAUGGGCUUGUUAGCUGUGCAAAUGUUUUUUUCUGCGAUUCAGGUCA-AACACUUGACCAGGUCGCAUCUGGUCAAAUAGAUCUCGAGCAAAGCGAGCACCUAUAAAUUAGCGAUC .(((....)))(((((((..((((...((((((.((((((((((..(((((-(....)))))).)))))))...((((.....))))..))).)))))).)))).......))))))).. ( -34.00) >DroEre_CAF1 4695 119 + 1 UGCCCCUGGGCUCGUAAGCUGUGCAAAUGUUUAUUUCUGCGAUUCAGGUCAAGCCAUUUGACCAGGUUGCAUCUGGUCAAAUAGAACUC-GGCAACCCGAGCGCCUAUAAAGUAGCGAUC .(((....)))((((..((...)).....(((((....(((.(((.(((...(((((((((((((((...)))))))))))).(....)-))).))).))))))..)))))...)))).. ( -35.90) >DroYak_CAF1 10264 120 + 1 UGGCAAUGGGCUUCUAAGCUGUGCAAAUGUUUAUUUCUGCGAUUCAGGUCAAGACAUUUGACCAGGUUGCAUCUGAUCAAAUAGAACUAAAGCAACCCGAGCUCCUAUAAAUUUGCGAUC ..((((.((((((......((..(((((((((...((((.....))))...)))))))))..))((((((....(.((.....)).)....)))))).)))).)).......)))).... ( -28.60) >consensus UGCCCAUGGGCUUGUAAGCUGUGCAAAUGUUUAUUUCUGCGAUUCAGGUCAAAACAUUUGACCAGGUUGCAUCUGGUCAAAUAGAUCUCGAGCAAAGCGAGCACCUAUAAAUUAGCGAUC .......((((((((..(((.................(((((((..((((((.....)))))).)))))))((((......)))).....)))...)))))).))............... (-25.42 = -25.70 + 0.28)

| Location | 1,554,454 – 1,554,574 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -27.60 |
| Energy contribution | -28.04 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1554454 120 - 20766785 GAUCGUUAAUUUAUAGGUGCCCGCUUUGCUCGAGAUCUAUUUGACCAGAUGCAACCUGGUCAAAUGUUUUGACCUGAAUCGCAGAAAUAAACAUUUGCACAGCUUACAAGCCCAUGGGCA .................(((((((((.(((((((((..((((((((((.......)))))))))))))))))........(((((........)))))...))....))))....))))) ( -33.00) >DroSec_CAF1 4277 120 - 1 GAUCGCUAAUUUAUAUGUGCUCGCUUUGCUCGAGAUCUAUUUGACCAGAUGCGACCUGGUCAAGUGUUUUGACCUGAAUCGCAGAAAAAAACAUUUGCACAGCUAACAAGCCCAUGGGCA ((.(((..........))).))(((.((((((((((..((((((((((.......))))))))))))))))).(((.....)))............))).)))......(((....))). ( -34.40) >DroSim_CAF1 4763 119 - 1 GAUCGCUAAUUUAUAGGUGCUCGCUUUGCUCGAGAUCUAUUUGACCAGAUGCGACCUGGUCAAGUGUU-UGACCUGAAUCGCAGAAAAAAACAUUUGCACAGCUAACAAGCCCAUGGGCA ....(((......(((((.((((.......))))...(((((((((((.......)))))))))))..-..)))))....(((((........)))))..)))......(((....))). ( -34.80) >DroEre_CAF1 4695 119 - 1 GAUCGCUACUUUAUAGGCGCUCGGGUUGCC-GAGUUCUAUUUGACCAGAUGCAACCUGGUCAAAUGGCUUGACCUGAAUCGCAGAAAUAAACAUUUGCACAGCUUACGAGCCCAGGGGCA ....(((......(((((((((((....))-))))..(((((((((((.......)))))))))))))))).((((....(((((........)))))...(((....))).))))))). ( -42.40) >DroYak_CAF1 10264 120 - 1 GAUCGCAAAUUUAUAGGAGCUCGGGUUGCUUUAGUUCUAUUUGAUCAGAUGCAACCUGGUCAAAUGUCUUGACCUGAAUCGCAGAAAUAAACAUUUGCACAGCUUAGAAGCCCAUUGCCA ....((((........(((((((((((.....((...(((((((((((.......))))))))))).)).)))))))...(((((........)))))...)))).........)))).. ( -30.23) >consensus GAUCGCUAAUUUAUAGGUGCUCGCUUUGCUCGAGAUCUAUUUGACCAGAUGCAACCUGGUCAAAUGUUUUGACCUGAAUCGCAGAAAUAAACAUUUGCACAGCUUACAAGCCCAUGGGCA ....(((......((((.((.......))((((((..(((((((((((.......)))))))))))))))))))))....(((((........)))))..)))......(((....))). (-27.60 = -28.04 + 0.44)

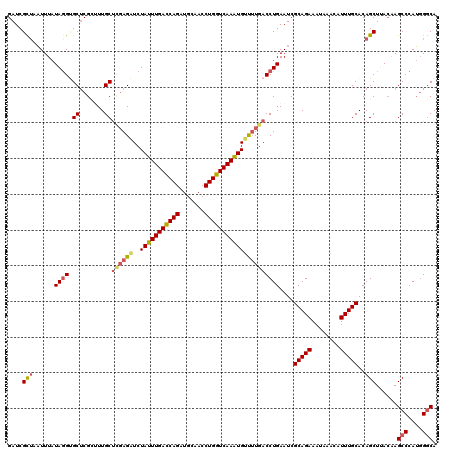
| Location | 1,554,494 – 1,554,614 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.17 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -25.64 |
| Energy contribution | -26.08 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.978015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1554494 120 + 20766785 GAUUCAGGUCAAAACAUUUGACCAGGUUGCAUCUGGUCAAAUAGAUCUCGAGCAAAGCGGGCACCUAUAAAUUAACGAUCAAUCCGAAAGCUUGCGAACUUUAACUCGCAACCAUGGAUA .(((((((((((.....)))))).((((((.((((......))))((.(((((....((((.....................))))...))))).))..........)))))).))))). ( -32.30) >DroSec_CAF1 4317 120 + 1 GAUUCAGGUCAAAACACUUGACCAGGUCGCAUCUGGUCAAAUAGAUCUCGAGCAAAGCGAGCACAUAUAAAUUAGCGAUCAAUGCGACCACUUGCGAACUUUAACUUGCAAUUAUGGAUA .(((((((((((.....)))))).((((((((.(((((.........(((.......)))((............)))))))))))))))..((((((........))))))...))))). ( -35.50) >DroSim_CAF1 4803 119 + 1 GAUUCAGGUCA-AACACUUGACCAGGUCGCAUCUGGUCAAAUAGAUCUCGAGCAAAGCGAGCACCUAUAAAUUAGCGAUCAAUGCGACCACUUGCGAACUUUAACUCGCAAUCAUGGAUA .((((((((((-(....)))))).((((((((.(((((........((((.......))))...(((.....))).)))))))))))))..((((((........))))))...))))). ( -39.30) >DroEre_CAF1 4735 119 + 1 GAUUCAGGUCAAGCCAUUUGACCAGGUUGCAUCUGGUCAAAUAGAACUC-GGCAACCCGAGCGCCUAUAAAGUAGCGAUCAUUGCGACAGCUUGCGAACUUUAACUCGCAACCAUGGAUA .((((((((.((((.((((((((((((...)))))))))))).(..(((-((....)))))..).......((.((((...)))).)).))))((((........)))).))).))))). ( -37.00) >DroYak_CAF1 10304 120 + 1 GAUUCAGGUCAAGACAUUUGACCAGGUUGCAUCUGAUCAAAUAGAACUAAAGCAACCCGAGCUCCUAUAAAUUUGCGAUCAAUCCGACAGCUUGCGAACUUUAAUUUGCAACUAUGAAUA .((((((((((((...))))))).(((((((((((......)))).....(((.......)))..........)))))))........((.(((((((......))))))))).))))). ( -25.80) >consensus GAUUCAGGUCAAAACAUUUGACCAGGUUGCAUCUGGUCAAAUAGAUCUCGAGCAAAGCGAGCACCUAUAAAUUAGCGAUCAAUGCGACAGCUUGCGAACUUUAACUCGCAACCAUGGAUA .(((((((((.......((((((((((...))))))))))((((..((((.......))))...))))........))))...........((((((........))))))...))))). (-25.64 = -26.08 + 0.44)

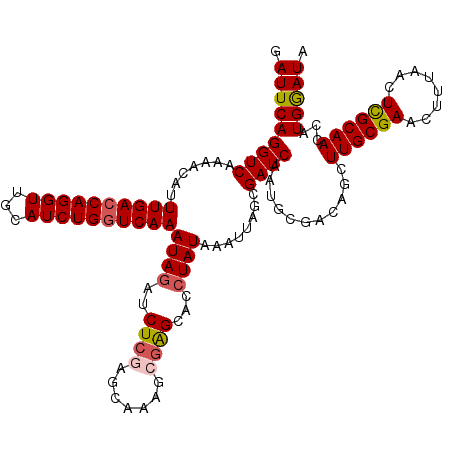
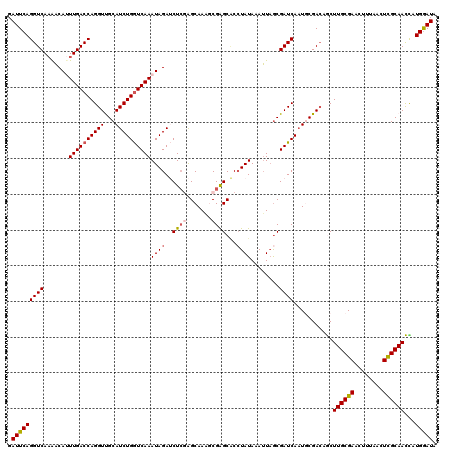
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:46 2006