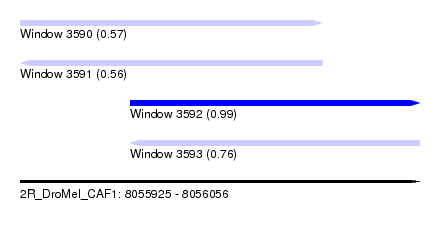
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,055,925 – 8,056,056 |
| Length | 131 |
| Max. P | 0.992008 |

| Location | 8,055,925 – 8,056,024 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -37.27 |
| Consensus MFE | -21.98 |
| Energy contribution | -23.26 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
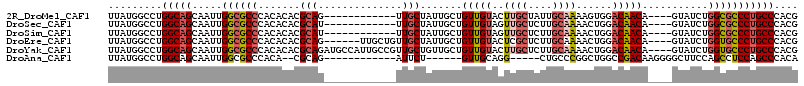
>2R_DroMel_CAF1 8055925 99 + 20766785 CGUGGGCAGGGCGCCAGAUAC----UGUUGUCCACUUUUGCAAUAGCAAGUACAACAGCAAUAGCAA------------CUGCGUGUGUGGGCGCCAAUUGCUGCCAGGCCAUAA .((.((((((((((((.((((----((((((.....(((((....)))))....))))))...((..------------..)))))).).)))))).....)))))..))..... ( -35.60) >DroSec_CAF1 20864 99 + 1 CGUGGGCAGGGCGCCAGAUAC----UGUUGUCCAGUUUUGCAAGAGCAACUACAACAGCAAUAGCAA------------AUGCGUGUGUGGGCGCCAAUUGCUGCCAGGCCAUAA .((.((((((((((((.((((----((((((..(((..(((....))))))))))))(((.......------------.))))))).).)))))).....)))))..))..... ( -35.00) >DroSim_CAF1 4773 99 + 1 CGUGGGCAGGGCGCCAGAUAC----UGUUGUCCAGUUUUGCAAGAGCAACUACAACAGCAAUAGCAA------------AUGCGUGUGUGGGCGCCAAUUGCUGCCAGGCCAUAA .((.((((((((((((.((((----((((((..(((..(((....))))))))))))(((.......------------.))))))).).)))))).....)))))..))..... ( -35.00) >DroEre_CAF1 19114 105 + 1 CGUGGGCAGGGCACCAGAUAC----UGUUGUCCAGUUUUGCAAGAGCGAGUACAACAGCAAUAGCAACAGCAA------CUGCGUGUGUGGGCGCCAAUUGCUGCCAGGCCAUAA ....(((..(((((((.((((----((((((.....(((((....))))).))))))(((...((....))..------.))))))).)))(((.....)))))))..))).... ( -34.90) >DroYak_CAF1 19049 111 + 1 CGUGGGCAGGGCACCAGAUAC----UGUUGUCCAGUUUUGCAAGAGCAAGUACAACAGCAACAGCAACGGCAAUGGCAUCUGCGUGUGUGGGCGCCAAUUGCUGCCAGGCCAUAA ....(((..(((........(----((((((...(((((((....))))).))....)))))))....(((((((((..(..(....)..)..))).)))))))))..))).... ( -40.50) >DroAna_CAF1 21258 90 + 1 UGUGGGCUGGAGGCUGGAAGCCCCUUGUCGGCCAGCCGGGCAG-----CCUGCAAC------AGAAU------------CUGCG--UGUGGGCGCCAAUUGCUGCCAGGCCAUAA (((((.((((.((((((..((.....))...)))))).(((.(-----((..((.(------((...------------)))..--))..))))))........)))).))))). ( -42.60) >consensus CGUGGGCAGGGCGCCAGAUAC____UGUUGUCCAGUUUUGCAAGAGCAACUACAACAGCAAUAGCAA____________CUGCGUGUGUGGGCGCCAAUUGCUGCCAGGCCAUAA ....(((..(((((((.((((....((((((......((((....)))).....))))))...(((..............))))))).)))(((.....)))))))..))).... (-21.98 = -23.26 + 1.28)


| Location | 8,055,925 – 8,056,024 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -20.53 |
| Energy contribution | -21.26 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8055925 99 - 20766785 UUAUGGCCUGGCAGCAAUUGGCGCCCACACACGCAG------------UUGCUAUUGCUGUUGUACUUGCUAUUGCAAAAGUGGACAACA----GUAUCUGGCGCCCUGCCCACG .........(((((.....((((((.......((..------------..))...(((((((((((((((....)))..)))..))))))----)))...))))))))))).... ( -34.80) >DroSec_CAF1 20864 99 - 1 UUAUGGCCUGGCAGCAAUUGGCGCCCACACACGCAU------------UUGCUAUUGCUGUUGUAGUUGCUCUUGCAAAACUGGACAACA----GUAUCUGGCGCCCUGCCCACG .........(((((.....((((((.......((..------------..))...(((((((((..((((....))))......))))))----)))...))))))))))).... ( -35.50) >DroSim_CAF1 4773 99 - 1 UUAUGGCCUGGCAGCAAUUGGCGCCCACACACGCAU------------UUGCUAUUGCUGUUGUAGUUGCUCUUGCAAAACUGGACAACA----GUAUCUGGCGCCCUGCCCACG .........(((((.....((((((.......((..------------..))...(((((((((..((((....))))......))))))----)))...))))))))))).... ( -35.50) >DroEre_CAF1 19114 105 - 1 UUAUGGCCUGGCAGCAAUUGGCGCCCACACACGCAG------UUGCUGUUGCUAUUGCUGUUGUACUCGCUCUUGCAAAACUGGACAACA----GUAUCUGGUGCCCUGCCCACG ....(((..(((((((((((.((........)))))------)))))...((((.(((((((((....((....))........))))))----)))..)))))))..))).... ( -33.90) >DroYak_CAF1 19049 111 - 1 UUAUGGCCUGGCAGCAAUUGGCGCCCACACACGCAGAUGCCAUUGCCGUUGCUGUUGCUGUUGUACUUGCUCUUGCAAAACUGGACAACA----GUAUCUGGUGCCCUGCCCACG .........(((((.....((((((...(((.((((..((....))..)))))))(((((((((..((((....))))......))))))----)))...))))))))))).... ( -36.80) >DroAna_CAF1 21258 90 - 1 UUAUGGCCUGGCAGCAAUUGGCGCCCACA--CGCAG------------AUUCU------GUUGCAGG-----CUGCCCGGCUGGCCGACAAGGGGCUUCCAGCCUCCAGCCCACA ....((((((.((((((((.(((......--))).)------------))..)------))))))))-----))....((((((((.....))(((.....))).)))))).... ( -35.90) >consensus UUAUGGCCUGGCAGCAAUUGGCGCCCACACACGCAG____________UUGCUAUUGCUGUUGUACUUGCUCUUGCAAAACUGGACAACA____GUAUCUGGCGCCCUGCCCACG .........(((((.....((((((.......(((..............))).......(((((..((((....))))......)))))...........))))))))))).... (-20.53 = -21.26 + 0.73)

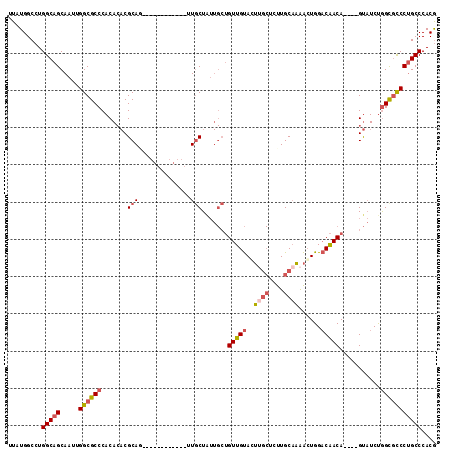
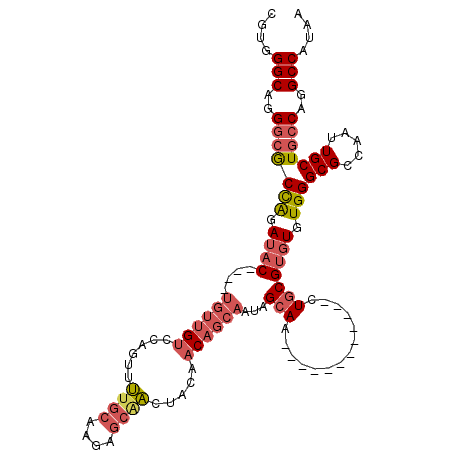
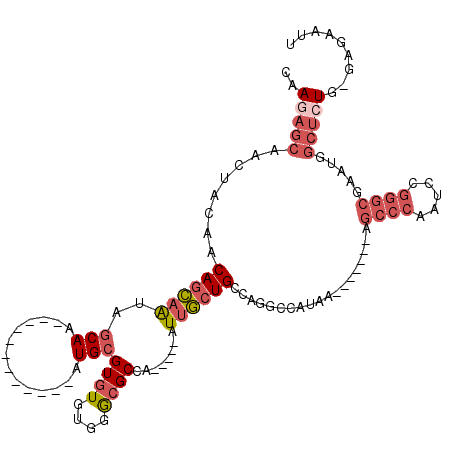
| Location | 8,055,961 – 8,056,056 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.49 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -16.38 |
| Energy contribution | -17.99 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8055961 95 + 20766785 CAAUAGCAAGUACAACAGCAAUAGCAA------------CUGCGUGUGUGGGCGCCA-----AUUGCUGCCAGGCCAUAA-------AGCCCAAUCCGGGCGAAUGGCUCUG-GAGAAUU ...............(((((((.((..------------(..(....)..)..))..-----)))))))(((((((((..-------.((((.....))))..))))).)))-)...... ( -35.10) >DroVir_CAF1 3119 98 + 1 CAACAGCAGCCGCAACAUUCGGGCCAA------------AUGCGUGUGGGCAGGCCAUAAAAAUAAUUGACAAAACAAAAAAUAACAAACUUAAUG----CGA------CUGCAAAAAUU .....((((.((((.......((((..------------.(((......)))))))..........(((......)))................))----)).------))))....... ( -22.00) >DroSec_CAF1 20900 95 + 1 CAAGAGCAACUACAACAGCAAUAGCAA------------AUGCGUGUGUGGGCGCCA-----AUUGCUGCCAGGCCAUAA-------AGCCCAAUCCGGGCGAAUGGCUCUG-GAGAAUU (((..((..(((((...(((.......------------.)))...)))))..))..-----.)))((.(((((((((..-------.((((.....))))..))))).)))-))).... ( -34.80) >DroSim_CAF1 4809 95 + 1 CAAGAGCAACUACAACAGCAAUAGCAA------------AUGCGUGUGUGGGCGCCA-----AUUGCUGCCAGGCCAUAA-------AGCCCAAUCCGGGCGAAUGGCUCUG-GAGAAUU (((..((..(((((...(((.......------------.)))...)))))..))..-----.)))((.(((((((((..-------.((((.....))))..))))).)))-))).... ( -34.80) >DroEre_CAF1 19150 101 + 1 CAAGAGCGAGUACAACAGCAAUAGCAACAGCAA------CUGCGUGUGUGGGCGCCA-----AUUGCUGCCAGGCCAUAA-------AGCCCAAUCCGGGCGAAUGGCUCUG-GAGAAUU (((..(((..((((...(((...((....))..------.)))...))))..)))..-----.)))((.(((((((((..-------.((((.....))))..))))).)))-))).... ( -37.10) >DroYak_CAF1 19085 107 + 1 CAAGAGCAAGUACAACAGCAACAGCAACGGCAAUGGCAUCUGCGUGUGUGGGCGCCA-----AUUGCUGCCAGGCCAUAA-------AGCCCAAUCCGGGCGAAUGGCUCUG-GAGAAUU .................((....))..((((((((((..(..(....)..)..))).-----)))))))(((((((((..-------.((((.....))))..))))).)))-)...... ( -40.80) >consensus CAAGAGCAACUACAACAGCAAUAGCAA____________AUGCGUGUGUGGGCGCCA_____AUUGCUGCCAGGCCAUAA_______AGCCCAAUCCGGGCGAAUGGCUCUG_GAGAAUU ..(((((........((((((..(((..............)))((((....))))........))))))...................((((.....)))).....)))))......... (-16.38 = -17.99 + 1.61)

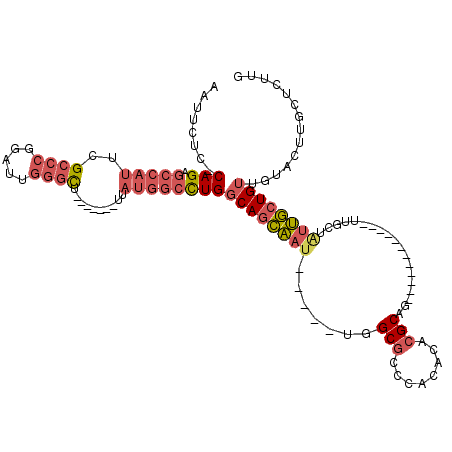
| Location | 8,055,961 – 8,056,056 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.49 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -16.44 |
| Energy contribution | -17.67 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8055961 95 - 20766785 AAUUCUC-CAGAGCCAUUCGCCCGGAUUGGGCU-------UUAUGGCCUGGCAGCAAU-----UGGCGCCCACACACGCAG------------UUGCUAUUGCUGUUGUACUUGCUAUUG (((((((-(((.(((((..((((.....)))).-------..))))))))).)).)))-----)((((...(((((.((((------------(....))))))).)))...)))).... ( -32.20) >DroVir_CAF1 3119 98 - 1 AAUUUUUGCAG------UCG----CAUUAAGUUUGUUAUUUUUUGUUUUGUCAAUUAUUUUUAUGGCCUGCCCACACGCAU------------UUGGCCCGAAUGUUGCGGCUGCUGUUG .......((((------(((----((....(((((.......(((......)))..........(((((((......))).------------..)))))))))..)))))))))..... ( -26.90) >DroSec_CAF1 20900 95 - 1 AAUUCUC-CAGAGCCAUUCGCCCGGAUUGGGCU-------UUAUGGCCUGGCAGCAAU-----UGGCGCCCACACACGCAU------------UUGCUAUUGCUGUUGUAGUUGCUCUUG ......(-(((.(((((..((((.....)))).-------..))))))))).((((((-----(((((........)))..------------..((....)).....)))))))).... ( -31.90) >DroSim_CAF1 4809 95 - 1 AAUUCUC-CAGAGCCAUUCGCCCGGAUUGGGCU-------UUAUGGCCUGGCAGCAAU-----UGGCGCCCACACACGCAU------------UUGCUAUUGCUGUUGUAGUUGCUCUUG ......(-(((.(((((..((((.....)))).-------..))))))))).((((((-----(((((........)))..------------..((....)).....)))))))).... ( -31.90) >DroEre_CAF1 19150 101 - 1 AAUUCUC-CAGAGCCAUUCGCCCGGAUUGGGCU-------UUAUGGCCUGGCAGCAAU-----UGGCGCCCACACACGCAG------UUGCUGUUGCUAUUGCUGUUGUACUCGCUCUUG (((((((-(((.(((((..((((.....)))).-------..))))))))).)).)))-----)((((...(((((.((((------(.((....)).))))))).)))...)))).... ( -36.80) >DroYak_CAF1 19085 107 - 1 AAUUCUC-CAGAGCCAUUCGCCCGGAUUGGGCU-------UUAUGGCCUGGCAGCAAU-----UGGCGCCCACACACGCAGAUGCCAUUGCCGUUGCUGUUGCUGUUGUACUUGCUCUUG ......(-(((.(((((..((((.....)))).-------..)))))))))(((((((-----.(((((........)).(....)...)))))))))).(((....))).......... ( -33.40) >consensus AAUUCUC_CAGAGCCAUUCGCCCGGAUUGGGCU_______UUAUGGCCUGGCAGCAAU_____UGGCGCCCACACACGCAG____________UUGCUAUUGCUGUUGUACUUGCUCUUG ........(((.(((((..((((.....))))..........))))))))((((((((.......(((........)))...................)))))))).............. (-16.44 = -17.67 + 1.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:07 2006