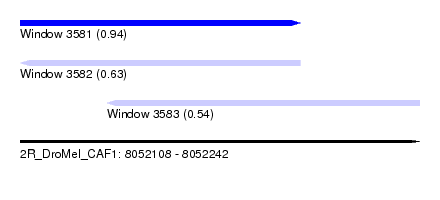
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,052,108 – 8,052,242 |
| Length | 134 |
| Max. P | 0.941488 |

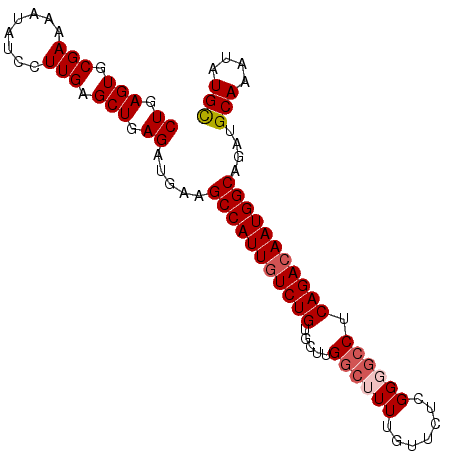
| Location | 8,052,108 – 8,052,202 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.53 |
| Mean single sequence MFE | -28.94 |
| Consensus MFE | -25.48 |
| Energy contribution | -26.32 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8052108 94 + 20766785 CUGAGUGCGAAAAUAUCCUUGAGCUGAGAUGAAGCCAUUGUCUGUGCUUGGCUUUUGUUCUCGGGGCCUCAGACAAUGGCAGAUGCAAAUAUGC ((.(((.(((........))).))).)).....(((((((((((.....((((((.......)))))).)))))))))))....(((....))) ( -29.90) >DroSec_CAF1 17117 94 + 1 CUGAGUGCGAAAAUAUCCUUGAGCUGAGAUGAAGCCAUUGUCUGUGCUUGGCUUUUGUUCUCGGGGCCUCAGACAAUGGCGGAUGCAAAUAUGU ((.(((.(((........))).))).)).....(((((((((((.....((((((.......)))))).))))))))))).............. ( -29.70) >DroSim_CAF1 313 94 + 1 CUGAGUGCGAAAAUAUCCUUGAGCUGAGAUGAAGCCAUUGUCUGUGCUUGGCUUUUGUUCUCGGGGCCUCAGACAAUGGCAGAUGCAAAUAUGC ((.(((.(((........))).))).)).....(((((((((((.....((((((.......)))))).)))))))))))....(((....))) ( -29.90) >DroEre_CAF1 15397 93 + 1 CUGAGUACGAAAAUAUCCUUGAGCUGAGAUGAAGCCAUUGUCUGUUCCUGGCUUUUGUUCUCGG-GCCUCAGACAAUGGCAGAUGCAAAUAUGC ((.(((.(((........))).))).)).....(((((((((((.....(((((........))-))).)))))))))))....(((....))) ( -29.70) >DroYak_CAF1 15261 93 + 1 CUGAGUGCGAAAAUAUCCUUGAGCUGAGUUGAAGCCAUUUUCUGUGCUUGGCUUUUGUUCUCGG-AACUCAGACAAUGGCAGAUGCAAAUAUGC ((((((.(((.((((.....((((..(((.((((....))))...)))..)))).)))).))).-.))))))......((....))........ ( -25.50) >consensus CUGAGUGCGAAAAUAUCCUUGAGCUGAGAUGAAGCCAUUGUCUGUGCUUGGCUUUUGUUCUCGGGGCCUCAGACAAUGGCAGAUGCAAAUAUGC ((.(((.(((........))).))).)).....(((((((((((.....((((((.......)))))).)))))))))))....(((....))) (-25.48 = -26.32 + 0.84)

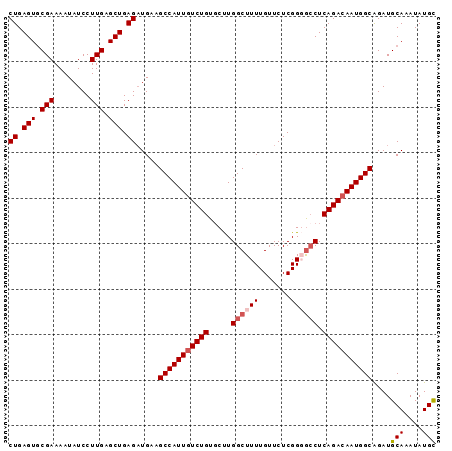
| Location | 8,052,108 – 8,052,202 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.53 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.18 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8052108 94 - 20766785 GCAUAUUUGCAUCUGCCAUUGUCUGAGGCCCCGAGAACAAAAGCCAAGCACAGACAAUGGCUUCAUCUCAGCUCAAGGAUAUUUUCGCACUCAG .......(((....(((((((((((.(((.............))).....)))))))))))...((((........))))......)))..... ( -23.62) >DroSec_CAF1 17117 94 - 1 ACAUAUUUGCAUCCGCCAUUGUCUGAGGCCCCGAGAACAAAAGCCAAGCACAGACAAUGGCUUCAUCUCAGCUCAAGGAUAUUUUCGCACUCAG .......(((....(((((((((((.(((.............))).....)))))))))))...((((........))))......)))..... ( -23.92) >DroSim_CAF1 313 94 - 1 GCAUAUUUGCAUCUGCCAUUGUCUGAGGCCCCGAGAACAAAAGCCAAGCACAGACAAUGGCUUCAUCUCAGCUCAAGGAUAUUUUCGCACUCAG .......(((....(((((((((((.(((.............))).....)))))))))))...((((........))))......)))..... ( -23.62) >DroEre_CAF1 15397 93 - 1 GCAUAUUUGCAUCUGCCAUUGUCUGAGGC-CCGAGAACAAAAGCCAGGAACAGACAAUGGCUUCAUCUCAGCUCAAGGAUAUUUUCGUACUCAG (((....)))....(((((((((((.(((-............))).....)))))))))))...((((........)))).............. ( -23.30) >DroYak_CAF1 15261 93 - 1 GCAUAUUUGCAUCUGCCAUUGUCUGAGUU-CCGAGAACAAAAGCCAAGCACAGAAAAUGGCUUCAACUCAGCUCAAGGAUAUUUUCGCACUCAG (((....)))...(((...((((((((((-..(((.....((((((...........))))))...))))))))..))))).....)))..... ( -18.90) >consensus GCAUAUUUGCAUCUGCCAUUGUCUGAGGCCCCGAGAACAAAAGCCAAGCACAGACAAUGGCUUCAUCUCAGCUCAAGGAUAUUUUCGCACUCAG .......(((....(((((((((((.(((.............))).....)))))))))))...............(((....))))))..... (-20.10 = -20.18 + 0.08)

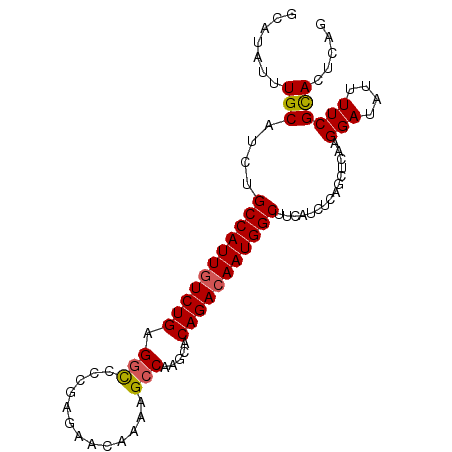
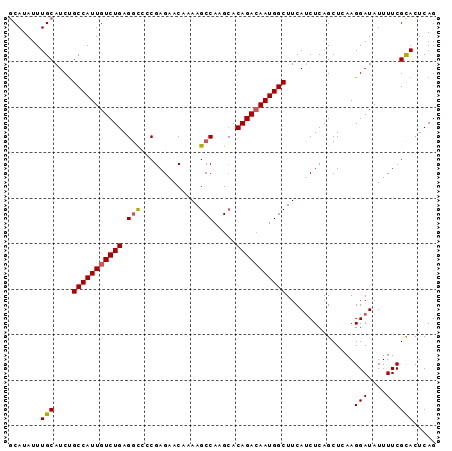
| Location | 8,052,137 – 8,052,242 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.69 |
| Mean single sequence MFE | -28.41 |
| Consensus MFE | -20.58 |
| Energy contribution | -21.50 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8052137 105 - 20766785 AGCGAGAGAACUUGUAUUUUACCACCUCAUUACAAGAUUUGCAUAUUUGCAUCUGCCAUUGUCUGAGGCC-CCGAGAACAAAAGCCAAGCACAGACAAUGGCUUCA-------------- ...((((((.((((((..............))))))...((((....)))))))(((((((((((.(((.-............))).....)))))))))))))).-------------- ( -29.56) >DroSec_CAF1 17146 105 - 1 AGCGAGAGAACUUGUAUUUUACCACCUCAUUACAAGAUUUACAUAUUUGCAUCCGCCAUUGUCUGAGGCC-CCGAGAACAAAAGCCAAGCACAGACAAUGGCUUCA-------------- .((((((((.((((((..............)))))).))).....)))))....(((((((((((.(((.-............))).....)))))))))))....-------------- ( -27.36) >DroSim_CAF1 342 105 - 1 AGCGAGAGAACUUGUAUUUUACCACCUCAUUACAAGAUUUGCAUAUUUGCAUCUGCCAUUGUCUGAGGCC-CCGAGAACAAAAGCCAAGCACAGACAAUGGCUUCA-------------- ...((((((.((((((..............))))))...((((....)))))))(((((((((((.(((.-............))).....)))))))))))))).-------------- ( -29.56) >DroEre_CAF1 15426 104 - 1 AGCGAGAGAACUUGUAUUUUACCACCUCAUUACAAGAUUUGCAUAUUUGCAUCUGCCAUUGUCUGAGGC--CCGAGAACAAAAGCCAGGAACAGACAAUGGCUUCA-------------- ...((((((.((((((..............))))))...((((....)))))))(((((((((((.(((--............))).....)))))))))))))).-------------- ( -29.64) >DroYak_CAF1 15290 104 - 1 AGCGAGAGAACUUGUAUUUUACCACCUCAUUACAAGAUUUGCAUAUUUGCAUCUGCCAUUGUCUGAGUU--CCGAGAACAAAAGCCAAGCACAGAAAAUGGCUUCA-------------- ...((((((.((((((..............))))))...((((....)))))))((((((.((((.(((--(...))))....((...)).)))).))))))))).-------------- ( -24.94) >DroAna_CAF1 18562 117 - 1 A---CGACAACUUGUACCUUACCACCUCAUUACUACAUUUGCAUAUUUGCAUCUGCCAUUGUCUGUGGCAGCCGAACAAAGAGGUGCAACUCAGACAAUGACAACAAUAGCCUGGUUGGU .---((((...(((((((((...................((((....)))).(((((((.....))))))).........)))))))))..(((.(.((.......)).).))))))).. ( -29.40) >consensus AGCGAGAGAACUUGUAUUUUACCACCUCAUUACAAGAUUUGCAUAUUUGCAUCUGCCAUUGUCUGAGGCC_CCGAGAACAAAAGCCAAGCACAGACAAUGGCUUCA______________ ..........((((((..............))))))...((((....))))...(((((((((((.(((..............))).....))))))))))).................. (-20.58 = -21.50 + 0.92)

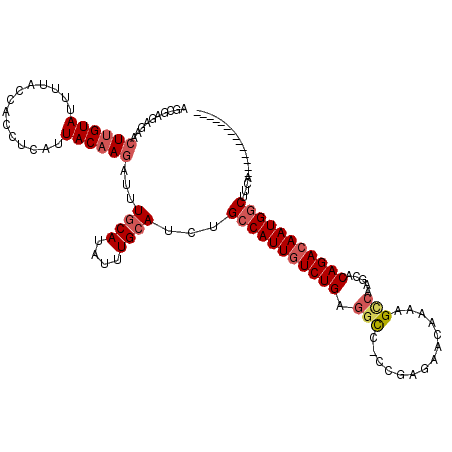
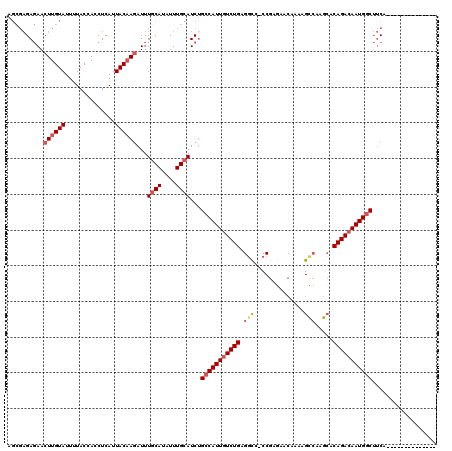
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:58 2006