| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,039,767 – 8,039,897 |
| Length | 130 |
| Max. P | 0.874724 |

| Location | 8,039,767 – 8,039,867 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.61 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -13.03 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
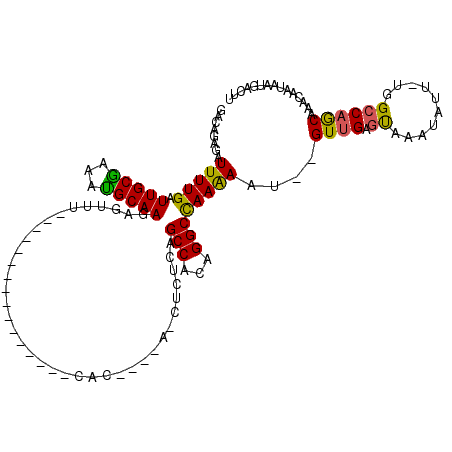
>2R_DroMel_CAF1 8039767 100 + 20766785 CCAUAACAUUCUCUUCCA--UUUCGCUUUUCAUUUGGAGCAAGUCAUUGCUGUUUGCUGGCCA-AAUGUUUACUCAAC--AUUUUUGGCCUGUGGCUGAGA--U----GUG--------- .....((((.(((.....--....(((((......))))).((((((.((.....)).(((((-((((((.....)))--))..)))))).))))))))))--)----)).--------- ( -27.80) >DroSec_CAF1 4779 100 + 1 CCACAACAUUCUCUUCCA--UUUCGCUUUUCAUUUCGAGCAAGUCAUUGCUGUUUGCUGGCCA-AAUAUUUAUUCAAC--AUUUUUGGCCUGUGGCUGAGA--U----GCG--------- ................((--((((((((........)))).((((((.((.....)).(((((-((............--...))))))).))))))))))--)----)..--------- ( -24.16) >DroEre_CAF1 3416 102 + 1 CAACAGCAUUCUCUUCCA--UUUCGUUUUCCAUUUAGAGCAAGUCAUUAUUGUUUGUUGGCUA-AAUAUUUACUCAAC--AUUUUUGGCCUGUGGCUGAGAGAU----GUG--------- .....((((((((..(((--(................((((((.((....))))))))(((((-((............--...))))))).))))..)))).))----)).--------- ( -22.46) >DroYak_CAF1 3056 106 + 1 CAACAACAUUCUCCUCCA--UUUCGUUUUCCAUUUAGAGCAAGUCAUUAUUGUUUGCUGGCCA-AAUAUUUACUCAAC--AUUUUUGGCCUGUGGCUGAAAGAAACCAUUG--------- ..................--((((.(((.((((....((((((.((....))))))))(((((-((............--...))))))).))))..))).))))......--------- ( -20.76) >DroAna_CAF1 5506 116 + 1 CAAUACCAUCUUCCUCCUGGCAUCUACUACCAUUUAGUGCCAGUCAUUAGUGUUUGUUGGCAAAAAUAUUUGCUCGACACACUCUUAGCCUUUGGCUGUGGGCU----GUGACUGUGGCU .....((((.........((((.(((........)))))))((((((.(((((..(((((((((....))))).)))))))))...(((((........)))))----)))))))))).. ( -33.70) >consensus CAACAACAUUCUCUUCCA__UUUCGCUUUCCAUUUAGAGCAAGUCAUUACUGUUUGCUGGCCA_AAUAUUUACUCAAC__AUUUUUGGCCUGUGGCUGAGAG_U____GUG_________ .........((((..((....................((((((.((....))))))))(((((.((................)).)))))...))..))))................... (-13.03 = -13.43 + 0.40)


| Location | 8,039,767 – 8,039,867 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.61 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -10.30 |
| Energy contribution | -11.50 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8039767 100 - 20766785 ---------CAC----A--UCUCAGCCACAGGCCAAAAAU--GUUGAGUAAACAUU-UGGCCAGCAAACAGCAAUGACUUGCUCCAAAUGAAAAGCGAAA--UGGAAGAGAAUGUUAUGG ---------.((----(--((((..(((..((((((..((--(((.....))))))-)))))(((((.((....))..))))).................--)))..))).))))..... ( -26.80) >DroSec_CAF1 4779 100 - 1 ---------CGC----A--UCUCAGCCACAGGCCAAAAAU--GUUGAAUAAAUAUU-UGGCCAGCAAACAGCAAUGACUUGCUCGAAAUGAAAAGCGAAA--UGGAAGAGAAUGUUGUGG ---------(((----(--((((..(((..((((((..((--(((.....))))))-))))).((((.((....))..))))(((...(....).)))..--)))..))))....)))). ( -26.40) >DroEre_CAF1 3416 102 - 1 ---------CAC----AUCUCUCAGCCACAGGCCAAAAAU--GUUGAGUAAAUAUU-UAGCCAACAAACAAUAAUGACUUGCUCUAAAUGGAAAACGAAA--UGGAAGAGAAUGCUGUUG ---------(((----((.((((..(((..(((...((((--(((.....))))))-).)))....................(((....)))........--)))..))))))).))... ( -16.00) >DroYak_CAF1 3056 106 - 1 ---------CAAUGGUUUCUUUCAGCCACAGGCCAAAAAU--GUUGAGUAAAUAUU-UGGCCAGCAAACAAUAAUGACUUGCUCUAAAUGGAAAACGAAA--UGGAGGAGAAUGUUGUUG ---------((((..(((((((((.(((..((((((..((--(((.....))))))-)))))(((((...........))))).....))).........--)))))))))..))))... ( -25.80) >DroAna_CAF1 5506 116 - 1 AGCCACAGUCAC----AGCCCACAGCCAAAGGCUAAGAGUGUGUCGAGCAAAUAUUUUUGCCAACAAACACUAAUGACUGGCACUAAAUGGUAGUAGAUGCCAGGAGGAAGAUGGUAUUG .((((((((((.----((((..........))))...(((((((.(.(((((....)))))).))..)))))..))))))...((...(((((.....)))))..)).....)))).... ( -32.40) >consensus _________CAC____A_CUCUCAGCCACAGGCCAAAAAU__GUUGAGUAAAUAUU_UGGCCAGCAAACAAUAAUGACUUGCUCUAAAUGGAAAACGAAA__UGGAAGAGAAUGUUGUUG ...................((((..((...(((((.......(((.....)))....)))))(((((.((....))..)))))....................))..))))......... (-10.30 = -11.50 + 1.20)

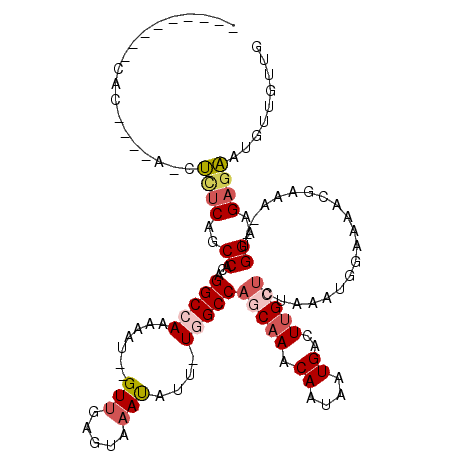
| Location | 8,039,805 – 8,039,897 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -9.18 |
| Energy contribution | -8.46 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8039805 92 - 20766785 ---AGAGAUUUUGAUUGCGAAACGCAAGAGUUU----------------CAC----A--UCUCAGCCACAGGCCAAAAAU--GUUGAGUAAACAUU-UGGCCAGCAAACAGCAAUGACUU ---.......(((.(((((((((......))))----------------)..----.--...........((((((..((--(((.....))))))-))))).)))).)))......... ( -23.90) >DroSec_CAF1 4817 92 - 1 ---ACAGAUUUUGAUUGCGAAACGCAAGAGUUU----------------CGC----A--UCUCAGCCACAGGCCAAAAAU--GUUGAAUAAAUAUU-UGGCCAGCAAACAGCAAUGACUU ---.......((((.((((((((......))))----------------)))----)--..)))).....((((((..((--(((.....))))))-))))).((.....))........ ( -25.70) >DroEre_CAF1 3454 97 - 1 GACAGAGAUUUUGAUUGCGAAAUGCAAGAGUCU----------------CAC----AUCUCUCAGCCACAGGCCAAAAAU--GUUGAGUAAAUAUU-UAGCCAACAAACAAUAAUGACUU ....(((((((...((((.....))))))))))----------------)..----..............(((...((((--(((.....))))))-).))).................. ( -17.00) >DroYak_CAF1 3094 101 - 1 GACAGAGAUUUUGAUUGCGAAAUGCAAGAGUUU----------------CAAUGGUUUCUUUCAGCCACAGGCCAAAAAU--GUUGAGUAAAUAUU-UGGCCAGCAAACAAUAAUGACUU ..........(((.(((((((((......))))----------------)..(((((......)))))..((((((..((--(((.....))))))-))))).)))).)))......... ( -23.30) >DroAna_CAF1 5546 116 - 1 GACAGAAAUUUUGAUUGCAAAAUGCAAGAGGACCAGUCCCAGCCACAGUCAC----AGCCCACAGCCAAAGGCUAAGAGUGUGUCGAGCAAAUAUUUUUGCCAACAAACACUAAUGACUG ..............((((.....))))(.(((....)))).....((((((.----((((..........))))...(((((((.(.(((((....)))))).))..)))))..)))))) ( -26.40) >consensus GACAGAGAUUUUGAUUGCGAAAUGCAAGAGUUU________________CAC____A_CUCUCAGCCACAGGCCAAAAAU__GUUGAGUAAAUAUU_UGGCCAGCAAACAAUAAUGACUU ........(((((.(((((...))))).....................................(((...))))))))....((((.((..........))))))............... ( -9.18 = -8.46 + -0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:41 2006