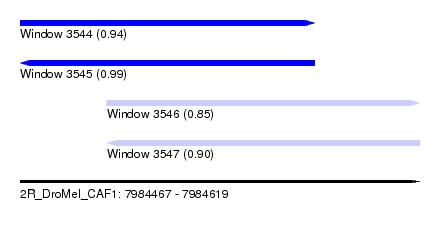
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,984,467 – 7,984,619 |
| Length | 152 |
| Max. P | 0.985716 |

| Location | 7,984,467 – 7,984,579 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.80 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -16.69 |
| Energy contribution | -16.95 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7984467 112 + 20766785 AUGUUGAAAAAAAAA-----CAUUGUUCAUAGCUAGUUAAAAGCUGAAAAGUUGACAGCACUGGCUCAAGCAAUUCCCU-CUGACCAGUGCUGGAAAACACUCGCUGUCUUUUGAGCG (((((........))-----))).(((((((((.(((....((((....))))..(((((((((.(((((.......))-.))))))))))))......))).)))).....))))). ( -32.10) >DroSim_CAF1 21339 111 + 1 AUAUUGAAAA-AAAA-----CAUUUUUCAUAGCUAGUUAAAAGCUAAAAAGUUGACAGCACUGGCUCAAGCAAUUCCCU-CUGACCAGUGCUGGAAAACACUCGCUGUCUUUUGAGCG ....((((((-(...-----..)))))))(((((.......)))))....(((..(((((((((.(((((.......))-.))))))))))))...)))...((((........)))) ( -30.80) >DroYak_CAF1 20964 111 + 1 AUUUUGGAUAGAAAA-----CAUAGUUCCCAGGUAGUUAUA-GCUAAAAAGUCGCCAGCACUGGCUUAAGCAGUUCCCU-GUGACCAGUGCUGGAAAACACUCGCUGUCUUUUGAGCG ...............-----....((((..((..(((....-(((....)))..((((((((((.....((((....))-))..)))))))))).........)))..))...)))). ( -30.80) >DroAna_CAF1 20838 103 + 1 GUACAGAUAA-AUAGUGACUGAUUUUUAUUA------------U--UGUAGUUGGCAACACCAACUUAAGAGAUUCCCAGCUGUGUAGUGUUGAAAAACACCCGGUGUCUUUUGAGCG .........(-(((((((.......))))))------------)--)((.(((((.....)))))(((((((((.....((((.(..(((((....))))))))))))))))))))). ( -21.70) >consensus AUAUUGAAAA_AAAA_____CAUUGUUCAUAGCUAGUUAAA_GCUAAAAAGUUGACAGCACUGGCUCAAGCAAUUCCCU_CUGACCAGUGCUGGAAAACACUCGCUGUCUUUUGAGCG ..................................................(((..(((((((((.(((.............))))))))))))...)))...((((........)))) (-16.69 = -16.95 + 0.25)

| Location | 7,984,467 – 7,984,579 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.80 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -19.23 |
| Energy contribution | -18.85 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7984467 112 - 20766785 CGCUCAAAAGACAGCGAGUGUUUUCCAGCACUGGUCAG-AGGGAAUUGCUUGAGCCAGUGCUGUCAACUUUUCAGCUUUUAACUAGCUAUGAACAAUG-----UUUUUUUUUCAACAU .....((((((((.....(((((..((((((((((..(-((.......)))..))))))))))..........((((.......))))..))))).))-----))))))......... ( -30.70) >DroSim_CAF1 21339 111 - 1 CGCUCAAAAGACAGCGAGUGUUUUCCAGCACUGGUCAG-AGGGAAUUGCUUGAGCCAGUGCUGUCAACUUUUUAGCUUUUAACUAGCUAUGAAAAAUG-----UUUU-UUUUCAAUAU (((((..........))))).....((((((((((..(-((.......)))..)))))))))).........(((((.......)))))(((((((..-----...)-)))))).... ( -32.70) >DroYak_CAF1 20964 111 - 1 CGCUCAAAAGACAGCGAGUGUUUUCCAGCACUGGUCAC-AGGGAACUGCUUAAGCCAGUGCUGGCGACUUUUUAGC-UAUAACUACCUGGGAACUAUG-----UUUUCUAUCCAAAAU .(((.(((((...(((((....)))((((((((((..(-((....))).....))))))))))))..))))).)))-...........(..(((...)-----))..).......... ( -32.70) >DroAna_CAF1 20838 103 - 1 CGCUCAAAAGACACCGGGUGUUUUUCAACACUACACAGCUGGGAAUCUCUUAAGUUGGUGUUGCCAACUACA--A------------UAAUAAAAAUCAGUCACUAU-UUAUCUGUAC .........(((.((((.(((.............))).))))..........((((((.....))))))...--.------------............))).....-.......... ( -18.72) >consensus CGCUCAAAAGACAGCGAGUGUUUUCCAGCACUGGUCAG_AGGGAAUUGCUUAAGCCAGUGCUGUCAACUUUUUAGC_UUUAACUAGCUAUGAAAAAUG_____UUUU_UUUUCAAUAU (((((..........))))).....((((((((((....(((......)))..))))))))))....................................................... (-19.23 = -18.85 + -0.38)


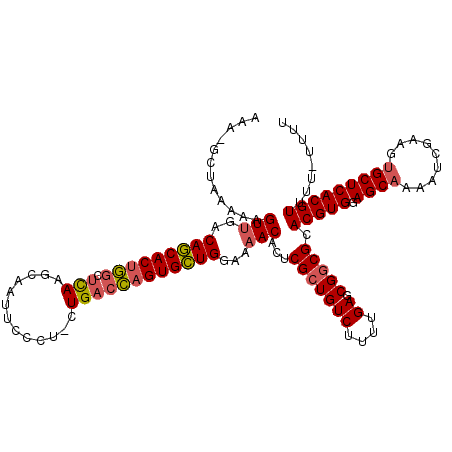
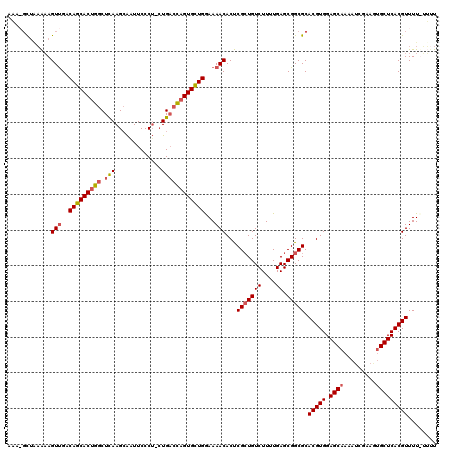
| Location | 7,984,500 – 7,984,619 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -28.69 |
| Energy contribution | -29.19 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7984500 119 + 20766785 AAAAGCUGAAAAGUUGACAGCACUGGCUCAAGCAAUUCCCU-CUGACCAGUGCUGGAAAACACUCGCUGUCUUUUGAGCGGCGCACGUGGAGCAAAAUCGAAGUGCUCACGUUUUUUUUU ...((((....))))..(((((((((.(((((.......))-.))))))))))))((((((...(((((((....)).)))))...(((.((((.........))))))))))))).... ( -38.10) >DroSim_CAF1 21371 117 + 1 AAAAGCUAAAAAGUUGACAGCACUGGCUCAAGCAAUUCCCU-CUGACCAGUGCUGGAAAACACUCGCUGUCUUUUGAGCGGCGCACGUGGAGCAAAAUCGAAGUGCUCACGUUUU--UUU ....((......(((..(((((((((.(((((.......))-.))))))))))))...)))....((((((....)).))))))(((((.((((.........)))))))))...--... ( -37.70) >DroYak_CAF1 20997 117 + 1 AUA-GCUAAAAAGUCGCCAGCACUGGCUUAAGCAGUUCCCU-GUGACCAGUGCUGGAAAACACUCGCUGUCUUUUGAGCGGCGCACGUGGAGCAAAAUCGAACUGCUCACGUUUU-UACU ...-((......((..((((((((((.....((((....))-))..))))))))))...))....((((((....)).))))))(((((.((((.........)))))))))...-.... ( -41.10) >DroAna_CAF1 20868 113 + 1 -----U--UGUAGUUGGCAACACCAACUUAAGAGAUUCCCAGCUGUGUAGUGUUGAAAAACACCCGGUGUCUUUUGAGCGGCGCACGUGGAGCAAAACAGAAGAGCUCACGUUUUUAGUC -----.--.((.(((((.....)))))(((((((((.....((((.(..(((((....)))))))))))))))))))))(((..(((((.(((...........)))))))).....))) ( -28.20) >consensus AAA_GCUAAAAAGUUGACAGCACUGGCUCAAGCAAUUCCCU_CUGACCAGUGCUGGAAAACACUCGCUGUCUUUUGAGCGGCGCACGUGGAGCAAAAUCGAAGUGCUCACGUUUU_UUUU ............(((..(((((((((.(((.............))))))))))))...)))...(((((((....)).))))).(((((.((((.........)))))))))........ (-28.69 = -29.19 + 0.50)

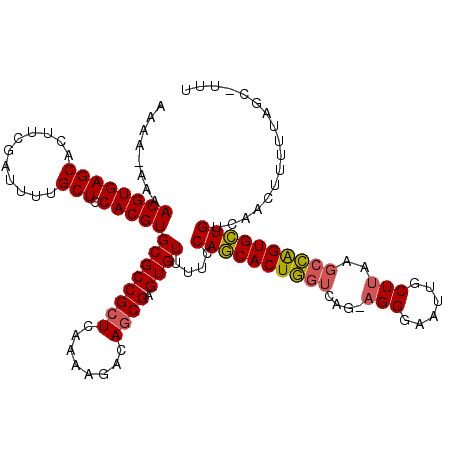
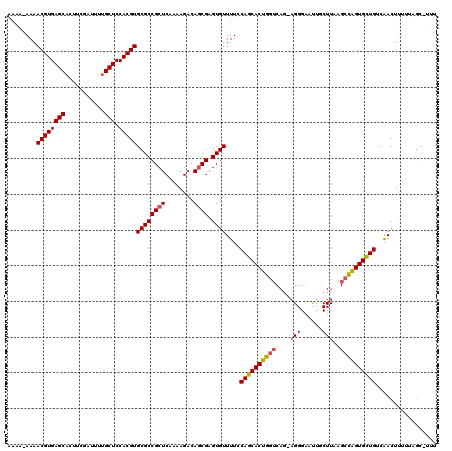
| Location | 7,984,500 – 7,984,619 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -36.41 |
| Consensus MFE | -30.49 |
| Energy contribution | -30.55 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7984500 119 - 20766785 AAAAAAAAACGUGAGCACUUCGAUUUUGCUCCACGUGCGCCGCUCAAAAGACAGCGAGUGUUUUCCAGCACUGGUCAG-AGGGAAUUGCUUGAGCCAGUGCUGUCAACUUUUCAGCUUUU ........(((((((((.........)))).)))))((((((((........)))).))))....((((((((((..(-((.......)))..))))))))))................. ( -37.90) >DroSim_CAF1 21371 117 - 1 AAA--AAAACGUGAGCACUUCGAUUUUGCUCCACGUGCGCCGCUCAAAAGACAGCGAGUGUUUUCCAGCACUGGUCAG-AGGGAAUUGCUUGAGCCAGUGCUGUCAACUUUUUAGCUUUU ...--...(((((((((.........)))).)))))((((((((........)))).))))....((((((((((..(-((.......)))..))))))))))................. ( -37.90) >DroYak_CAF1 20997 117 - 1 AGUA-AAAACGUGAGCAGUUCGAUUUUGCUCCACGUGCGCCGCUCAAAAGACAGCGAGUGUUUUCCAGCACUGGUCAC-AGGGAACUGCUUAAGCCAGUGCUGGCGACUUUUUAGC-UAU (((.-(((((((((((((.......))))).)))))((((((((........)))).))))...(((((((((((..(-((....))).....)))))))))))......))).))-).. ( -43.70) >DroAna_CAF1 20868 113 - 1 GACUAAAAACGUGAGCUCUUCUGUUUUGCUCCACGUGCGCCGCUCAAAAGACACCGGGUGUUUUUCAACACUACACAGCUGGGAAUCUCUUAAGUUGGUGUUGCCAACUACA--A----- ........((((((((...........))).)))))((((((((.((.(((..((((.(((.............))).))))...))).)).)).))))))...........--.----- ( -26.12) >consensus AAAA_AAAACGUGAGCACUUCGAUUUUGCUCCACGUGCGCCGCUCAAAAGACAGCGAGUGUUUUCCAGCACUGGUCAG_AGGGAAUUGCUUAAGCCAGUGCUGUCAACUUUUUAGC_UUU ........((((((((...........))).)))))((((((((........)))).))))....((((((((((....(((......)))..))))))))))................. (-30.49 = -30.55 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:22 2006