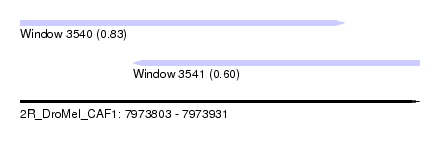
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,973,803 – 7,973,931 |
| Length | 128 |
| Max. P | 0.833064 |

| Location | 7,973,803 – 7,973,907 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -25.13 |
| Consensus MFE | -19.92 |
| Energy contribution | -20.37 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
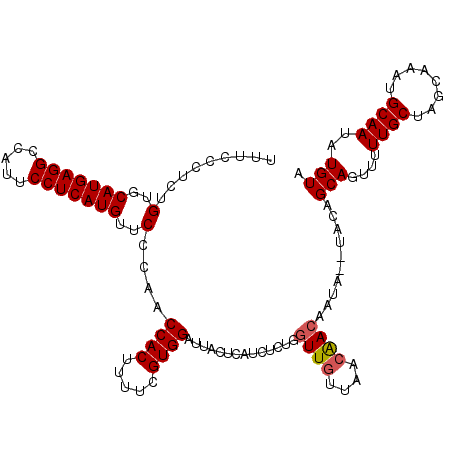
>2R_DroMel_CAF1 7973803 104 + 20766785 UUUCCAUGUGUCCAUGAGGCCAUUCCUCAUGUUCCUAGCCACUUUUCGUGGACU------------UUUUCAACGACAAUAUAUAUAGCAGUUUUUGCUUGCAAAUGCAAUAUGUA ........(((((((((((.....))))))).....((((((.....)))).))------------........)))).(((((((.(((...((((....))))))).))))))) ( -25.40) >DroSec_CAF1 10115 114 + 1 UUUCCCUCUGUGCAUGAGGCCAUUCCUCAUGUUCCCAACCACUUUUCGUGGAUUACUCAUCUCUGGUUGUUAACAACAAUA--UACAGCAGUUUUUGCUAGCAAAUGCAAUAUGUA .....((((((((((((((.....))))))))......((((.....))))..............((((....))))....--.)))).))...((((........))))...... ( -25.00) >DroSim_CAF1 10138 114 + 1 UUUCCCUCUGUGCAUGAGGCCAUUCCUCAUGUUCCCAACCACUUUUCGUGGAUUACUCAUCUCUGGUUGUUAACAACAAUA--UACAGCAGUUUUUGCUAGCAAAUGCAAUAUGUA .....((((((((((((((.....))))))))......((((.....))))..............((((....))))....--.)))).))...((((........))))...... ( -25.00) >consensus UUUCCCUCUGUGCAUGAGGCCAUUCCUCAUGUUCCCAACCACUUUUCGUGGAUUACUCAUCUCUGGUUGUUAACAACAAUA__UACAGCAGUUUUUGCUAGCAAAUGCAAUAUGUA .........(..(((((((.....)))))))..)....((((.....))))..............((((....))))..........(((....((((........))))..))). (-19.92 = -20.37 + 0.45)

| Location | 7,973,839 – 7,973,931 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 85.16 |
| Mean single sequence MFE | -17.33 |
| Consensus MFE | -15.47 |
| Energy contribution | -15.37 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
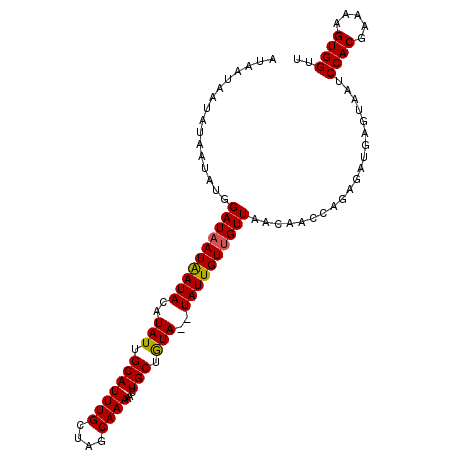
>2R_DroMel_CAF1 7973839 92 - 20766785 AUAAUAAUAUAACAUGGAUAAUGAUACAUAUUGCAUUUGCAAGCAAAAACUGCUAUAUAUAUUGUCGUUGAAAA------------AGUCCACGAAAAGUGGCU ..................((((((((.(((((((....)))((((.....))))....))))))))))))....------------((.((((.....)))))) ( -16.80) >DroSec_CAF1 10151 102 - 1 AUAAUAAUAUAAUAUGGAUAAUAAUACAUAUUGCAUUUGCUAGCAAAAACUGCUGUA--UAUUGUUGUUAACAACCAGAGAUGAGUAAUCCACGAAAAGUGGUU ((((((((((((((((..........)))))))....(((.((((.....)))))))--))))))))).....................((((.....)))).. ( -17.60) >DroSim_CAF1 10174 102 - 1 AUAAUAAUAUAAUAUGGAUAAUAAUACAUAUUGCAUUUGCUAGCAAAAACUGCUGUA--UAUUGUUGUUAACAACCAGAGAUGAGUAAUCCACGAAAAGUGGUU ((((((((((((((((..........)))))))....(((.((((.....)))))))--))))))))).....................((((.....)))).. ( -17.60) >consensus AUAAUAAUAUAAUAUGGAUAAUAAUACAUAUUGCAUUUGCUAGCAAAAACUGCUGUA__UAUUGUUGUUAACAACCAGAGAUGAGUAAUCCACGAAAAGUGGUU ................((((((((((..(((.(((((((....))))...))).)))..))))))))))....................((((.....)))).. (-15.47 = -15.37 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:17 2006