
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,966,059 – 7,966,168 |
| Length | 109 |
| Max. P | 0.780902 |

| Location | 7,966,059 – 7,966,168 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.71 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -23.84 |
| Energy contribution | -22.60 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
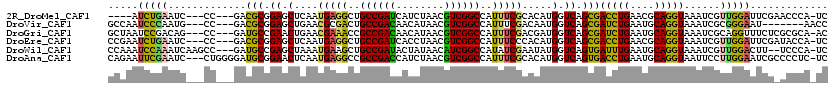
>2R_DroMel_CAF1 7966059 109 + 20766785 ----AUCUGAAUC---CC---GACGCGGAGCUCAAUGAGGCUGCCGAUCAUCUAACGUCGGCCAUUUCGCACAUGGUCAGCGACCUGAACGCAGGUAAAUCGUUGGAUUCGAACCCA-UC ----...((((((---(.---(((((((((((......)))).))).........(((.((((((.......)))))).)))(((((....)))))....)))))))))))......-.. ( -35.00) >DroVir_CAF1 2853 107 + 1 GCCAAUCCCAAUG---CC---GACGCGGAGCUGAACGCGACUGCCGACAACAUAACGUCGGCCAUUUCGACAAUGGUCAGCGAUCUGAAUGCAGGUAAAUCGCGGGAAU-------AACC .....((((...(---((---(.((((........))))...((((((........))))))...........))))..((((((((....)))....)))))))))..-------.... ( -34.00) >DroGri_CAF1 2874 113 + 1 GCUAAUCCGACAG---CC---GAUGCCGAACUGAACGAAACCGCCGACAACAUAACGUCGGCCAUUUCGACGAUGGUCAGCGAUCUGAAUGCAGGUAAAUCGCAGGUUUCUCGCGCA-AC (((........))---).---..((((((.......((((((((((((........))))))......(((....))).((((((((....)))....))))).))))))))).)))-.. ( -33.21) >DroEre_CAF1 2393 113 + 1 CCGAAUCUGAAUC---CC---GACGCGGAGCUCAAUGAGGCUGCCGAUCACCUAACGUCGGCCAUUUCCCACAUGGUCAGCGACCUGAACGCAGGUAAAUCGUUGGAUUCGAUACCA-UC ....(((.(((((---(.---(((((((((((......)))).))).........(((.((((((.......)))))).)))(((((....)))))....)))))))))))))....-.. ( -35.80) >DroWil_CAF1 20877 114 + 1 CCAAAUCCAAAUCAAGCC---GAUGCCGAGCUAAAUGAAGCUGCCGAUACUAUAACAUCGGCCAUAUCGAAUAUGGUCAGUGAUUUGAAUGCAGGUAAAUCGUUGGACUU--UCCCA-UC ((..((.(((((((.(((---(((..((((((......))).((((((........))))))....))).)).))))...))))))).))...))........(((....--..)))-.. ( -25.60) >DroAna_CAF1 3407 116 + 1 CAGAAUUCGAAUC---CUGGGGAUGCGGAACUCAAUGAGGCCGCCGACCAUCUAACGUCGGCCAUUUCGCACAUGGUCAGUGACCUGAAUGCAGGUAAUUCCUUGGAAUCGCCCCUC-UC .(((...(((.((---(.(((((((((((((((...)))...((((((........))))))..))))))............(((((....))))).)))))).))).)))....))-). ( -37.50) >consensus CCGAAUCCGAAUC___CC___GACGCGGAGCUCAAUGAGGCUGCCGACAACAUAACGUCGGCCAUUUCGAACAUGGUCAGCGACCUGAAUGCAGGUAAAUCGUUGGAUUCG_UCCCA_UC .....(((((.............(((.((.(....(((((..((((((........))))))..))))).....).)).)))(((((....)))))......)))))............. (-23.84 = -22.60 + -1.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:11 2006