
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
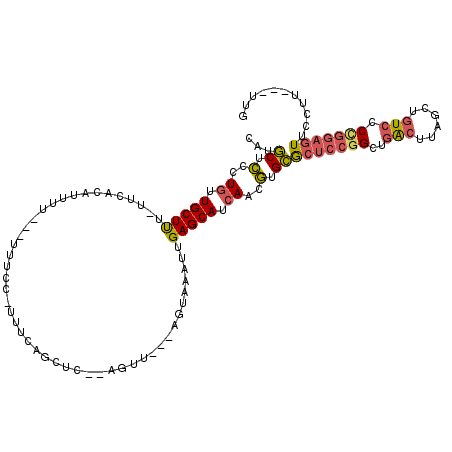
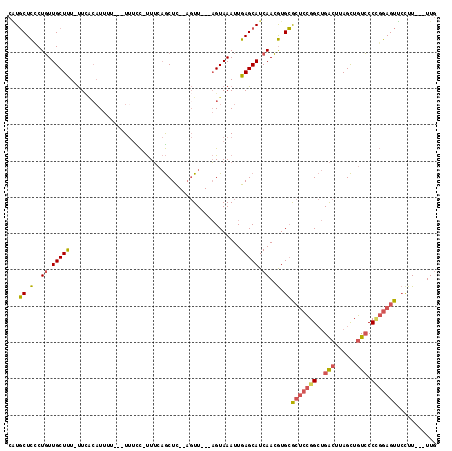
| Location | 7,960,616 – 7,960,724 |
| Length | 108 |
| Max. P | 0.758294 |

| Location | 7,960,616 – 7,960,724 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.64 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -14.19 |
| Energy contribution | -14.55 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7960616 108 + 20766785 CAUGCUCCCUGUUGCUUU-UUCACAUUUUUUUUUUCC-UUUCAGCUC--AGUU---AGUAAAUUGAGCAUCAACGUGCGCUCCGGCUGACUUAGCUGUCCCUGGAGUUCCUU---UUG ...((........))...-..............((((-...(((((.--((((---(((.....(.((((....)))).)....))))))).))))).....))))......---... ( -24.60) >DroSec_CAF1 12574 105 + 1 CAUGCUCCCUGUUGCUUU-UUAGCAUUUU---UUUCC-UUUCAGCUC--AGUU---AGUAAAUUGAGCAUCAACGUGCGCUCCGGCUGACUUAGCUGUCCCCGGAGUUCCUU---UUG ...((.(.....((((..-..))))....---.....-.....((((--((((---....))))))))......).))(((((((..(((......))).))))))).....---... ( -29.60) >DroSim_CAF1 12270 105 + 1 CAUGCUCGCUGUUGCUUU-UUCACAUUUU---UUUCC-UUUCAGCUC--AGUU---AGUAAAUUGAGCAUCAACGUGCGCUCCGGCUGACUUAGCUGUCCCCGGAGUUCCUU---UUG ...((.(((....))...-..........---.....-.....((((--((((---....))))))))......).))(((((((..(((......))).))))))).....---... ( -28.70) >DroEre_CAF1 12628 101 + 1 CGUGCUCCCUGUUGCUUU-UUCA--UU-----UUUUC-UUUCAGCUC--AGUU---AGUAAAUUGAGCAUCAACGUGCGCUCCGGCUGACUUAGCUGUCCCCGGAGUUCUUU---UUG (((((........))...-....--..-----.....-.....((((--((((---....))))))))....))).(.(((((((..(((......))).))))))).)...---... ( -26.90) >DroMoj_CAF1 18760 111 + 1 CUUUCUUGCUCUUGCUCUGCUUCCAU------UACUUUCUGC-ACUUCUUCUUUGGCGUAAAUUGAGCAUCAAGAUGAAAGCUGGAAGGAAAAGUUAUGCCAUAAGUAGCUUAAGUUC ((((((.(((((((...(((((...(------(((.....((-.(.........)))))))...))))).))))).....)).))))))..(((((((.......)))))))...... ( -19.70) >consensus CAUGCUCCCUGUUGCUUU_UUCACAUUUU___UUUCC_UUUCAGCUC__AGUU___AGUAAAUUGAGCAUCAACGUGCGCUCCGGCUGACUUAGCUGUCCCCGGAGUUCCUU___UUG ...((.(..((.(((((...............................................))))).))..).))(((((((..(((......))).)))))))........... (-14.19 = -14.55 + 0.36)

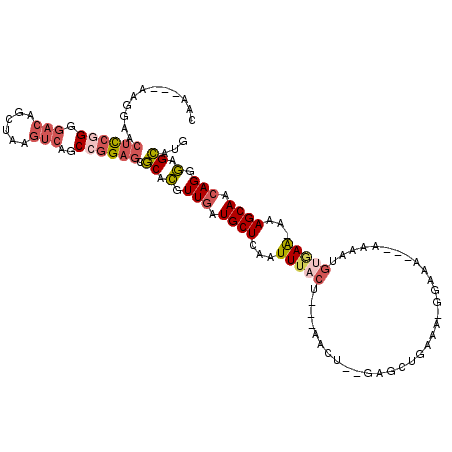
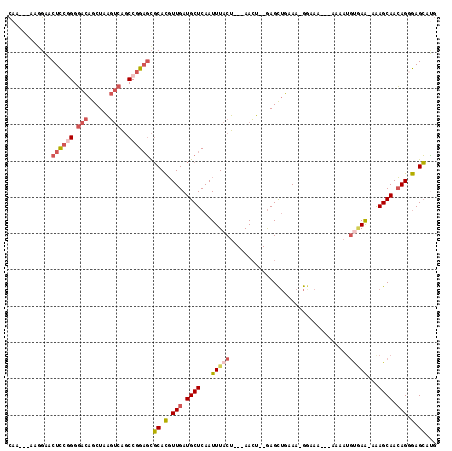
| Location | 7,960,616 – 7,960,724 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.64 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -13.63 |
| Energy contribution | -15.15 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7960616 108 - 20766785 CAA---AAGGAACUCCAGGGACAGCUAAGUCAGCCGGAGCGCACGUUGAUGCUCAAUUUACU---AACU--GAGCUGAAA-GGAAAAAAAAAAUGUGAA-AAAGCAACAGGGAGCAUG ...---......((((..(..(((((.(((.((...((((((.....).)))))......))---.)))--.)))))...-............(((...-...))).)..)))).... ( -22.90) >DroSec_CAF1 12574 105 - 1 CAA---AAGGAACUCCGGGGACAGCUAAGUCAGCCGGAGCGCACGUUGAUGCUCAAUUUACU---AACU--GAGCUGAAA-GGAAA---AAAAUGCUAA-AAAGCAACAGGGAGCAUG (((---...(..((((((.(((......)))..))))))..)...)))((((((........---..((--(..((....-))...---....((((..-..)))).))).)))))). ( -30.30) >DroSim_CAF1 12270 105 - 1 CAA---AAGGAACUCCGGGGACAGCUAAGUCAGCCGGAGCGCACGUUGAUGCUCAAUUUACU---AACU--GAGCUGAAA-GGAAA---AAAAUGUGAA-AAAGCAACAGCGAGCAUG ...---......((((((.(((......)))..)))))).((.(((((.((((...(((((.---....--...((....-))...---.....)))))-..)))).))))).))... ( -33.19) >DroEre_CAF1 12628 101 - 1 CAA---AAAGAACUCCGGGGACAGCUAAGUCAGCCGGAGCGCACGUUGAUGCUCAAUUUACU---AACU--GAGCUGAAA-GAAAA-----AA--UGAA-AAAGCAACAGGGAGCACG (((---...(..((((((.(((......)))..))))))..)...))).(((((........---..((--(.(((....-.....-----..--....-..)))..))).))))).. ( -24.43) >DroMoj_CAF1 18760 111 - 1 GAACUUAAGCUACUUAUGGCAUAACUUUUCCUUCCAGCUUUCAUCUUGAUGCUCAAUUUACGCCAAAGAAGAAGU-GCAGAAAGUA------AUGGAAGCAGAGCAAGAGCAAGAAAG ...(((..((((....))))....(((((.(((...(((((((((((..(((.(..(((........)))...).-)))..)))..------)))))))).))).))))).))).... ( -20.70) >consensus CAA___AAGGAACUCCGGGGACAGCUAAGUCAGCCGGAGCGCACGUUGAUGCUCAAUUUACU___AACU__GAGCUGAAA_GGAAA___AAAAUGUGAA_AAAGCAACAGGGAGCAUG ............((((((.(((......)))..)))))).((.(.(((.((((...(((((.................................)))))...)))).))).).))... (-13.63 = -15.15 + 1.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:06 2006