
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,949,620 – 7,949,750 |
| Length | 130 |
| Max. P | 0.985802 |

| Location | 7,949,620 – 7,949,735 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.77 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -21.22 |
| Energy contribution | -22.03 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7949620 115 + 20766785 CGUUAGUUCGUAGCGCAGCGAGUGAUGCAUCAGGGCAAUCUGCUGCUGCACUUUCAUGAGCAGCACUGAAUUGCACUCGAUAAGCAGGAUUUGCCUGCUAAAACUGCAAAUGGAA ......(((((...((((((((((.....(((((((.....)))(((((.(......).))))).))))....))))))...((((((.....))))))....))))..))))). ( -41.20) >DroVir_CAF1 1311 114 + 1 CGUCAGCUCGUAACGCAUCGAAUGAUGCAUCAACGCGAUCUGCUGCUGCACCUUCUGCAACAACAUCGAACCGCUCUCGAUGAGCAGGAUUUGCCGCACAAAGCUGCGGGGG-AG ..((..((((((..(((((....)))))......((....((.(((.(((..((((((.....((((((.......)))))).))))))..))).)))))..)))))))).)-). ( -36.50) >DroPse_CAF1 1285 115 + 1 UGUCAGCUCACACCGCAUCGAGUGGUGCAUAAGGGCAAUCUGUUGCUGCACCUUCAGCAGCAGCAAAGAAUCGCUCUGGACCAGCAGUAUCUGGCGGCUGAAACUGAGAGAGAAA ..((((((..((((((.....))))))....(((((..(((((((((((.......))))))))..)))...))))).(.((((......))))))))))).............. ( -39.30) >DroGri_CAF1 1342 109 + 1 CGUCAGCUCGUAACGCAUCGAAUGAUGCAUCAACGCAAUCUGCUGCUGCACCUUCAGCAGCAGCAACGAGCCGCUCUCAAUCAGUAGAAUCUGACGCACAAAACUGCAA------ .((((((((((...(((((....)))))............(((((((((.......)))))))))))))))...((((.....).)))...))))(((......)))..------ ( -38.80) >DroMoj_CAF1 1830 114 + 1 CGUCAGCUCGUAGCGCAGCGAGUGGUGCAUCAAUGCCAACUGCUGCUGCACCUUCAGCAGCAACAAGGAGCCGCUCUCAAUAAGCAGGAUUUGACGCACGAAGCUGCAGGGG-AA (.((.((.....))(((((...(.((((.((((..((...((((((((......))))))))..........(((.......))).))..)))).)))).).))))).)).)-.. ( -40.80) >DroPer_CAF1 1285 115 + 1 UGUCAGCUCACACCGCAUCGAGUGGUGCAUAAGGGCAAUCUGUUGCUGCACCUUCAGCAGCAGCAAAGAAUCGCUCUGGACCAGCAGUAUCUGGCGGCUGAAACUGAGAGGGAAA ..((((((..((((((.....))))))....(((((..(((((((((((.......))))))))..)))...))))).(.((((......)))))))))))..((....)).... ( -40.90) >consensus CGUCAGCUCGUAACGCAUCGAGUGAUGCAUCAAGGCAAUCUGCUGCUGCACCUUCAGCAGCAGCAAAGAACCGCUCUCGAUCAGCAGGAUCUGACGCACAAAACUGCAAGGG_AA ((((((.((.....(((((....)))))............(((((((((.......)))))))))..))...(((.......))).....))))))................... (-21.22 = -22.03 + 0.81)


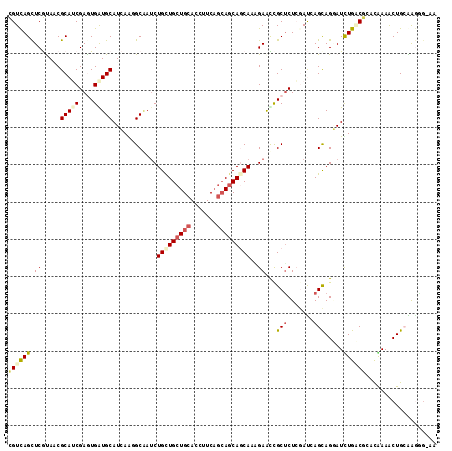
| Location | 7,949,620 – 7,949,735 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.77 |
| Mean single sequence MFE | -41.90 |
| Consensus MFE | -25.68 |
| Energy contribution | -25.88 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7949620 115 - 20766785 UUCCAUUUGCAGUUUUAGCAGGCAAAUCCUGCUUAUCGAGUGCAAUUCAGUGCUGCUCAUGAAAGUGCAGCAGCAGAUUGCCCUGAUGCAUCACUCGCUGCGCUACGAACUAACG ........(((((...((((((.....))))))....(((((....((((((((((((((....))).))))))).......)))).....)))))))))).............. ( -38.71) >DroVir_CAF1 1311 114 - 1 CU-CCCCCGCAGCUUUGUGCGGCAAAUCCUGCUCAUCGAGAGCGGUUCGAUGUUGUUGCAGAAGGUGCAGCAGCAGAUCGCGUUGAUGCAUCAUUCGAUGCGUUACGAGCUGACG ..-....((((((((...(((((.....((((((.....)))))).....((((((((((.....))))))))))).))))((.((((((((....)))))))))))))))).)) ( -45.40) >DroPse_CAF1 1285 115 - 1 UUUCUCUCUCAGUUUCAGCCGCCAGAUACUGCUGGUCCAGAGCGAUUCUUUGCUGCUGCUGAAGGUGCAGCAACAGAUUGCCCUUAUGCACCACUCGAUGCGGUGUGAGCUGACA ..............(((((((((((......)))))..((.((((((..(((((((.((.....)))))))))..)))))).))...(((((.(.....).)))))).))))).. ( -38.00) >DroGri_CAF1 1342 109 - 1 ------UUGCAGUUUUGUGCGUCAGAUUCUACUGAUUGAGAGCGGCUCGUUGCUGCUGCUGAAGGUGCAGCAGCAGAUUGCGUUGAUGCAUCAUUCGAUGCGUUACGAGCUGACG ------..(((......)))(((((......))))).....((((((((((((((((((.......)))))))))).......(((((((((....)))))))))))))))).). ( -45.50) >DroMoj_CAF1 1830 114 - 1 UU-CCCCUGCAGCUUCGUGCGUCAAAUCCUGCUUAUUGAGAGCGGCUCCUUGUUGCUGCUGAAGGUGCAGCAGCAGUUGGCAUUGAUGCACCACUCGCUGCGCUACGAGCUGACG ..-.....(((((...(((((((((....(((((.....)))))(((..((((((((((.......))))))))))..))).))))))))).....)))))((.....))..... ( -45.80) >DroPer_CAF1 1285 115 - 1 UUUCCCUCUCAGUUUCAGCCGCCAGAUACUGCUGGUCCAGAGCGAUUCUUUGCUGCUGCUGAAGGUGCAGCAACAGAUUGCCCUUAUGCACCACUCGAUGCGGUGUGAGCUGACA ..............(((((((((((......)))))..((.((((((..(((((((.((.....)))))))))..)))))).))...(((((.(.....).)))))).))))).. ( -38.00) >consensus UU_CCCUCGCAGUUUCAGCCGCCAAAUCCUGCUGAUCGAGAGCGAUUCGUUGCUGCUGCUGAAGGUGCAGCAGCAGAUUGCCCUGAUGCACCACUCGAUGCGCUACGAGCUGACG .........((((((...((.....................((((((...(((((((((.......)))))))))))))))......(((.(....).)))))...))))))... (-25.68 = -25.88 + 0.20)


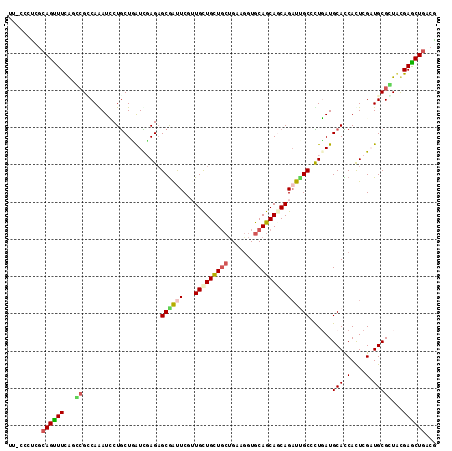
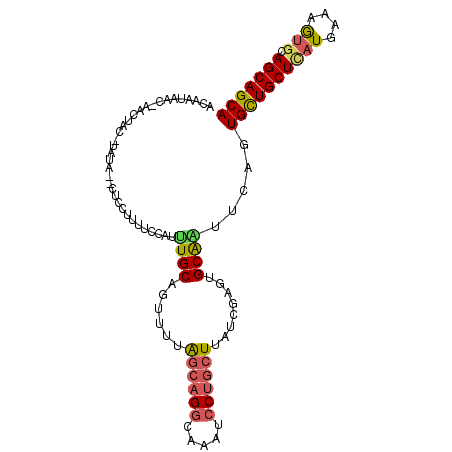
| Location | 7,949,660 – 7,949,750 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.58 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -15.35 |
| Energy contribution | -16.35 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7949660 90 + 20766785 UGCUGCUGCACUUUCAUGAGCAGCACUGAAUUGCACUCGAUAAGCAGGAUUUGCCUGCUAAAACUGCAAAUGGAAAAUGAG--UAUAUGUAG----------- (((((((.((......)))))))))(((...((.(((((...((((((.....))))))....((......))....))))--).))..)))----------- ( -23.90) >DroSec_CAF1 1409 101 + 1 UGCUGCUGCACUUUCAUGAGCAGCACUGAAUUGCCUUCGAUAAGCAGGAUUUGCCUGCUAAAACUGCAAAUGGAAUAGGAG--UAUAUGUAGUUUGUUAUUGU (((((((.((......)))))))))............(((((((((((.....)))))).((((((((.(((.........--))).))))))))..))))). ( -28.00) >DroSim_CAF1 1412 101 + 1 UGCUGCUGCACUUUCAUGAGCAGCACUGAAUUGCCUUCGAUAAGCAGGAUUUGCCUGCUAAAACUGCAAAUGGAAAAGGAG--UAUAUGUAGUUUGUUAUUGU (((((((.((......)))))))))............(((((((((((.....)))))).((((((((.(((.........--))).))))))))..))))). ( -28.00) >DroEre_CAF1 1434 97 + 1 UGCUGCUGCACUUUCAUAAGCAGCACUGAAUUGCACUCGAUAAGCAGGAUUUGCCUGCUAAAACUGCAUAAGGAAAAGUGG--U----GUAGUUAGUUAUUGU .((((((...........))))))(((((.(((((((.....((((((.....))))))....((.....)).......))--)----)))))))))...... ( -28.30) >DroYak_CAF1 1398 97 + 1 UGCUGCUGCAUUUUCAUGAGCAGCACUGAACUGCACUCGAUAAGCAGGAUUUGCCUGCUAAAACUGCAAAAGGAAAAUGGG--U----GCAGUUAGUUAUUAU .((((((.(((....)))))))))(((((.(((((((((...((((((.....))))))....((.....)).....))))--)----)))))))))...... ( -37.80) >DroMoj_CAF1 1870 91 + 1 UGCUGCUGCACCUUCAGCAGCAACAAGGAGCCGCUCUCAAUAAGCAGGAUUUGACGCACGAAGCUGCAGGGG-AAGAGGAGAUUAAA-G----------ACAG ((((((((......))))))))........((.(((((.....((((..((((.....)))).))))..)))-.)).))........-.----------.... ( -24.20) >consensus UGCUGCUGCACUUUCAUGAGCAGCACUGAAUUGCACUCGAUAAGCAGGAUUUGCCUGCUAAAACUGCAAAUGGAAAAGGAG__UAUA_GUAGUU_GUUAUUGU (((((((((..........)))))).................((((((.....))))))......)))................................... (-15.35 = -16.35 + 1.00)

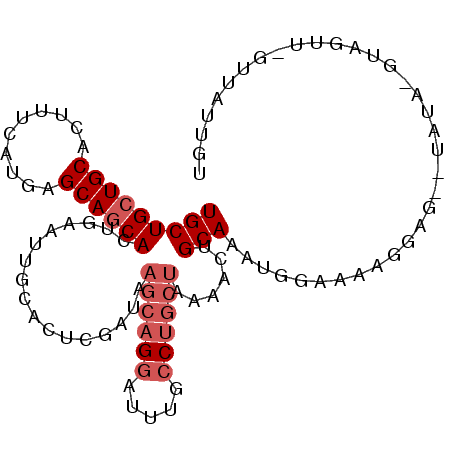
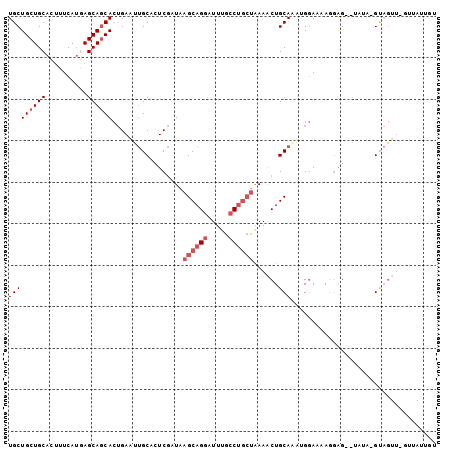
| Location | 7,949,660 – 7,949,750 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.58 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -20.33 |
| Energy contribution | -20.56 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7949660 90 - 20766785 -----------CUACAUAUA--CUCAUUUUCCAUUUGCAGUUUUAGCAGGCAAAUCCUGCUUAUCGAGUGCAAUUCAGUGCUGCUCAUGAAAGUGCAGCAGCA -----------.........--............(((((.((..((((((.....))))))....)).))))).....((((((((((....))).))))))) ( -28.90) >DroSec_CAF1 1409 101 - 1 ACAAUAACAAACUACAUAUA--CUCCUAUUCCAUUUGCAGUUUUAGCAGGCAAAUCCUGCUUAUCGAAGGCAAUUCAGUGCUGCUCAUGAAAGUGCAGCAGCA ....................--............((((..((..((((((.....))))))....))..)))).....((((((((((....))).))))))) ( -26.60) >DroSim_CAF1 1412 101 - 1 ACAAUAACAAACUACAUAUA--CUCCUUUUCCAUUUGCAGUUUUAGCAGGCAAAUCCUGCUUAUCGAAGGCAAUUCAGUGCUGCUCAUGAAAGUGCAGCAGCA ....................--............((((..((..((((((.....))))))....))..)))).....((((((((((....))).))))))) ( -26.60) >DroEre_CAF1 1434 97 - 1 ACAAUAACUAACUAC----A--CCACUUUUCCUUAUGCAGUUUUAGCAGGCAAAUCCUGCUUAUCGAGUGCAAUUCAGUGCUGCUUAUGAAAGUGCAGCAGCA ...............----.--.............((((.((..((((((.....))))))....)).))))......((((((((((....))).))))))) ( -26.10) >DroYak_CAF1 1398 97 - 1 AUAAUAACUAACUGC----A--CCCAUUUUCCUUUUGCAGUUUUAGCAGGCAAAUCCUGCUUAUCGAGUGCAGUUCAGUGCUGCUCAUGAAAAUGCAGCAGCA ......(((((((((----(--(.(......((.....))....((((((.....))))))....).)))))))).)))(((((((((....))).)))))). ( -33.50) >DroMoj_CAF1 1870 91 - 1 CUGU----------C-UUUAAUCUCCUCUU-CCCCUGCAGCUUCGUGCGUCAAAUCCUGCUUAUUGAGAGCGGCUCCUUGUUGCUGCUGAAGGUGCAGCAGCA ....----------.-..............-...(((((.(((((.(((.(((...((((((.....)))))).......))).)))))))).)))))..... ( -24.30) >consensus ACAAUAAC_AACUAC_UAUA__CUCCUUUUCCAUUUGCAGUUUUAGCAGGCAAAUCCUGCUUAUCGAGUGCAAUUCAGUGCUGCUCAUGAAAGUGCAGCAGCA ..................................((((......((((((.....))))))........)))).....((((((((((....))).))))))) (-20.33 = -20.56 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:00 2006