| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,949,260 – 7,949,447 |
| Length | 187 |
| Max. P | 0.846739 |

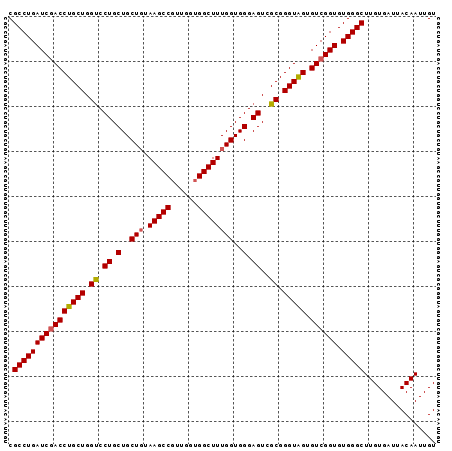
| Location | 7,949,260 – 7,949,355 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -35.51 |
| Energy contribution | -35.73 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7949260 95 + 20766785 CGCCUGAUCAACCUGCUGGCCCUGCUGCUUUAAGCCGUUGGUGGCUUUGGUGGGAGUCGCGGGUGGUGUCGGUGUGGGCUUGUGAUUACAAUUGU ((((((((..((((((.(((((..((.....((((((....)))))).))..)).)))))))))...))))).)))................... ( -34.90) >DroSec_CAF1 1012 92 + 1 CGCCUGAUCGACCUGCUGGUCCUGCUGCUGUAAGCCGUU---GGCUUUGGUGGGAGUCGCGGGUAGUGUCGGUGUGGGCUUGUGAUUACAAUUGU .((((((((((((((((.((.((.(..(((.(((((...---))))))))..).))..)).))))).)))))).)))))................ ( -36.20) >DroSim_CAF1 1012 95 + 1 CGCCUGAUCGACCUGCUGGUCCUGCUGCUGUAAGCCGUUGGUGGCUUUGGUGGGAGUCGCGGGUAGUGUCGGUGUGGGCUUGUGAUUACAAUUGU .((((((((((((((((.((.((.(..(((.((((((....)))))))))..).))..)).))))).)))))).)))))................ ( -38.10) >consensus CGCCUGAUCGACCUGCUGGUCCUGCUGCUGUAAGCCGUUGGUGGCUUUGGUGGGAGUCGCGGGUAGUGUCGGUGUGGGCUUGUGAUUACAAUUGU .((((((((((((((((.((.((.(..(((.(((((......))))))))..).))..)).))))).)))))).)))))................ (-35.51 = -35.73 + 0.23)


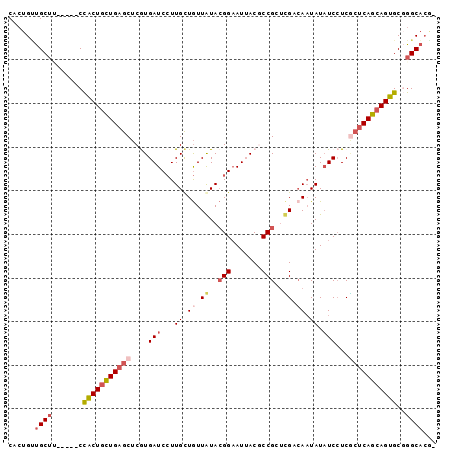
| Location | 7,949,355 – 7,949,447 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -18.65 |
| Energy contribution | -19.77 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7949355 92 + 20766785 CACUGUUGCUU-----CCACUGCUGAGCUCGUGAUCCUUGCUGUUGUACGGAAUAACGCCGCUCGACAAUAUAUCCUCGCUCAGCAGUGCGGGCACG- ......((((.-----.(((((((((((..(.(((...((.(((((..(((.......)))..))))).)).))))..)))))))))))..))))..- ( -37.10) >DroVir_CAF1 1041 95 + 1 C---GGCGCACUCGGCCUGCUAUUGUGCACAUGAUCAUUGCUAUUAUAGGGUAUAACGCCGGCGGCCACUAUCUCAUCCGUCAGCAGCGCCGGCUCGC (---(((((..(((...(((......)))..)))....((((.......(((.....)))(((((............)))))))))))))))...... ( -28.70) >DroSec_CAF1 1104 92 + 1 CACUGUUGCUU-----CCACUGCUGAGCUCGUGAUCCUUGCUGUUGUACGGAAUUACCCCGCUCGACAAUAUAUCCUCGCUCAGCAGUGCGGGCAAA- .....(((((.-----.(((((((((((..(.(((...((.(((((..(((.......)))..))))).)).))))..)))))))))))..))))).- ( -38.50) >DroSim_CAF1 1107 92 + 1 CACUGUUGCUU-----CCACUGCUGAGCUCGUGAUCCUUGCUGUUGUACGGAAUUACCCCGCUCGACAAUAUAUCCUCGCUCAGCAGUGCGGGCACA- ......((((.-----.(((((((((((..(.(((...((.(((((..(((.......)))..))))).)).))))..)))))))))))..))))..- ( -37.30) >DroEre_CAF1 1129 92 + 1 CACUGCUGCUA-----CCACUGCUGAGCUCAUGAUCCUUGUUGUUAUACGGAAUUACGCCGCUUGUGAAUAUAUCCUCACUCAGCAGUGCCGGCACG- ......((((.-----.((((((((((.......(((.(((....))).))).(((((.....)))))...........))))))))))..))))..- ( -26.60) >DroYak_CAF1 1093 92 + 1 CACUGCUGCUA-----CCACUGCUGAGCUCGUGAUCCUUGCUGUUAUACGGAAUUACGCCGCUUGUCAAUAUAUCCUCGCUCAGCAGUGCGGGCACG- ......((((.-----.(((((((((((..(.(((...((.((.((..(((.......)))..)).)).)).))))..)))))))))))..))))..- ( -29.70) >consensus CACUGUUGCUU_____CCACUGCUGAGCUCGUGAUCCUUGCUGUUAUACGGAAUUACGCCGCUCGACAAUAUAUCCUCGCUCAGCAGUGCGGGCACG_ ......((((.......(((((((((((....(((...((.((.((..(((.......)))..)).)).)).)))...)))))))))))..))))... (-18.65 = -19.77 + 1.11)


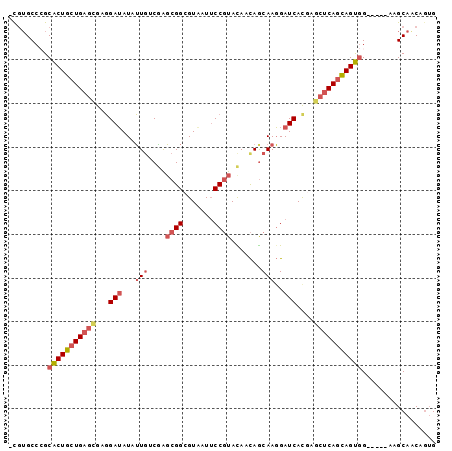
| Location | 7,949,355 – 7,949,447 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -33.29 |
| Consensus MFE | -21.58 |
| Energy contribution | -22.72 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7949355 92 - 20766785 -CGUGCCCGCACUGCUGAGCGAGGAUAUAUUGUCGAGCGGCGUUAUUCCGUACAACAGCAAGGAUCACGAGCUCAGCAGUGG-----AAGCAACAGUG -..(((...(((((((((((..((((...((((.(.((((.......)))).).)))).....))).)..))))))))))).-----..)))...... ( -33.60) >DroVir_CAF1 1041 95 - 1 GCGAGCCGGCGCUGCUGACGGAUGAGAUAGUGGCCGCCGGCGUUAUACCCUAUAAUAGCAAUGAUCAUGUGCACAAUAGCAGGCCGAGUGCGCC---G ..((((((((((..(((.(......).)))..).)))))))(((((........))))).....))..((((((.............)))))).---. ( -32.82) >DroSec_CAF1 1104 92 - 1 -UUUGCCCGCACUGCUGAGCGAGGAUAUAUUGUCGAGCGGGGUAAUUCCGUACAACAGCAAGGAUCACGAGCUCAGCAGUGG-----AAGCAACAGUG -.((((...(((((((((((..((((...((((.(.(((((.....))))).).)))).....))).)..))))))))))).-----..))))..... ( -35.50) >DroSim_CAF1 1107 92 - 1 -UGUGCCCGCACUGCUGAGCGAGGAUAUAUUGUCGAGCGGGGUAAUUCCGUACAACAGCAAGGAUCACGAGCUCAGCAGUGG-----AAGCAACAGUG -(((((...(((((((((((..((((...((((.(.(((((.....))))).).)))).....))).)..))))))))))).-----..)).)))... ( -35.20) >DroEre_CAF1 1129 92 - 1 -CGUGCCGGCACUGCUGAGUGAGGAUAUAUUCACAAGCGGCGUAAUUCCGUAUAACAACAAGGAUCAUGAGCUCAGCAGUGG-----UAGCAGCAGUG -..(((.(.((((((((((((((......)))))..((..(((...(((............)))..))).))))))))))).-----).)))...... ( -30.30) >DroYak_CAF1 1093 92 - 1 -CGUGCCCGCACUGCUGAGCGAGGAUAUAUUGACAAGCGGCGUAAUUCCGUAUAACAGCAAGGAUCACGAGCUCAGCAGUGG-----UAGCAGCAGUG -..(((...(((((((((((..((((...(((.......(((......))).......)))..))).)..))))))))))).-----..)))...... ( -32.34) >consensus _CGUGCCCGCACUGCUGAGCGAGGAUAUAUUGUCGAGCGGCGUAAUUCCGUACAACAGCAAGGAUCACGAGCUCAGCAGUGG_____AAGCAACAGUG .........(((((((((((...(((...(((....((((.......)))).......)))..)))....)))))))))))................. (-21.58 = -22.72 + 1.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:56 2006