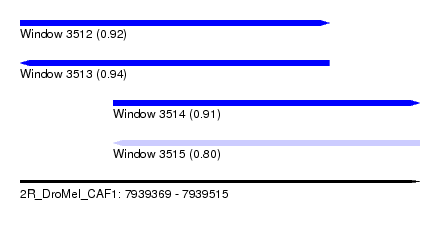
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,939,369 – 7,939,515 |
| Length | 146 |
| Max. P | 0.938503 |

| Location | 7,939,369 – 7,939,482 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -31.69 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7939369 113 + 20766785 UUUU-----AGCAAUUUUAAAUUUUAAAAUGCGGUUGCUCAAAAUAAUCUGGCAGCUCUUGCAACCUUAUGGCAUCCGUGCGAACAGCUGUCAGAUCAGCUGUUGUUGUUUCCAGCCC ....-----.(((.(((((.....))))))))(((((.........(((((((((((.(((((.((....))......)))))..)))))))))))((((....))))....))))). ( -32.60) >DroSec_CAF1 17839 99 + 1 -------------------AAUUUUAAAAUGCGGUUGCUCCAAAUAAUCUGGCAGCUCUUGUAACCUUCUGGCAACGGUGCGGACAGCUUUCAGAUCAGCUGUUGUUGCUGCCAGCCC -------------------.............((.....)).......((((((((........((..(((....)))...))((((((........))))))....))))))))... ( -29.30) >DroSim_CAF1 19387 113 + 1 UUUU-----AGCAAUUUCAAAUUUUAAAAUGCGGUUGCUCCAAAUAAUCUGGCAGCUCUUGUAACCUUCUGGCAACGGUGCGGACAGCUGUCAGAUCAGCUGUUGUUGCUGCCAGCCC ....-----((((((..((..........))..)))))).........((((((((........((..(((....)))...))(((((((......)))))))....))))))))... ( -37.30) >DroEre_CAF1 16269 118 + 1 UUAUAUAUUUGCAAUUUCAAAUUUAAAUAUGCGCUAGUUCCAAAUCAUCUGGCAGCUCUUGGAACCUUCUGGCAUCCGUGUGAACAGCUGUCAGAUCAGCUGUUCUUACUGCCAGCCC ...((((((((.(((.....))))))))))).....(((((((...............)))))))...((((((...(((.(((((((((......))))))))).)))))))))... ( -33.46) >DroYak_CAF1 23285 117 + 1 UGUAAUAUGUGCAAUUUCAAUUUUAAA-AUGCGGUUGAUCCAAAUUAUCUGGCAUCUCUUGGUACCGUCUGGCAUCCGUGCGAGCAGCUGUCAGAUCAGCUGUUUUUGUCGCUAGCCC ((((.....))))..............-..(((((.((((((.......))).))).......)))))..(((...((...(((((((((......)))))))))....))...))). ( -25.80) >consensus UUUU_____AGCAAUUUCAAAUUUUAAAAUGCGGUUGCUCCAAAUAAUCUGGCAGCUCUUGGAACCUUCUGGCAUCCGUGCGAACAGCUGUCAGAUCAGCUGUUGUUGCUGCCAGCCC ................................................((((((((...............(((....))).((((((((......))))))))...))))))))... (-23.34 = -23.50 + 0.16)

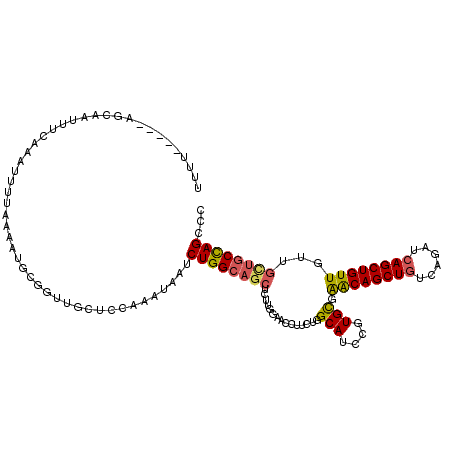
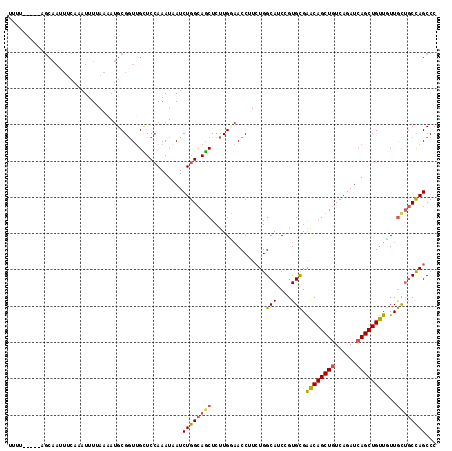
| Location | 7,939,369 – 7,939,482 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -21.46 |
| Energy contribution | -22.34 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7939369 113 - 20766785 GGGCUGGAAACAACAACAGCUGAUCUGACAGCUGUUCGCACGGAUGCCAUAAGGUUGCAAGAGCUGCCAGAUUAUUUUGAGCAACCGCAUUUUAAAAUUUAAAAUUGCU-----AAAA ((((((..........))))(((((((.(((((.((.(((.....(((....)))))))).))))).)))))))..........))((((((((.....))))).))).-----.... ( -31.20) >DroSec_CAF1 17839 99 - 1 GGGCUGGCAGCAACAACAGCUGAUCUGAAAGCUGUCCGCACCGUUGCCAGAAGGUUACAAGAGCUGCCAGAUUAUUUGGAGCAACCGCAUUUUAAAAUU------------------- ...((((((((....((((((........)))))).......((.(((....))).))....))))))))..........((....))...........------------------- ( -30.10) >DroSim_CAF1 19387 113 - 1 GGGCUGGCAGCAACAACAGCUGAUCUGACAGCUGUCCGCACCGUUGCCAGAAGGUUACAAGAGCUGCCAGAUUAUUUGGAGCAACCGCAUUUUAAAAUUUGAAAUUGCU-----AAAA ...((((((((....(((((((......))))))).......((.(((....))).))....)))))))).........(((((...((..........))...)))))-----.... ( -35.10) >DroEre_CAF1 16269 118 - 1 GGGCUGGCAGUAAGAACAGCUGAUCUGACAGCUGUUCACACGGAUGCCAGAAGGUUCCAAGAGCUGCCAGAUGAUUUGGAACUAGCGCAUAUUUAAAUUUGAAAUUGCAAAUAUAUAA ...((((((((..(((((((((......)))))))))....(((.(((....))))))....))))))))................(((..((((....))))..))).......... ( -38.60) >DroYak_CAF1 23285 117 - 1 GGGCUAGCGACAAAAACAGCUGAUCUGACAGCUGCUCGCACGGAUGCCAGACGGUACCAAGAGAUGCCAGAUAAUUUGGAUCAACCGCAU-UUUAAAAUUGAAAUUGCACAUAUUACA ((....((((......((((((......)))))).))))..((.((((....))))))....(((.(((((...))))))))..))((((-((((....))))).))).......... ( -29.10) >consensus GGGCUGGCAGCAACAACAGCUGAUCUGACAGCUGUUCGCACGGAUGCCAGAAGGUUACAAGAGCUGCCAGAUUAUUUGGAGCAACCGCAUUUUAAAAUUUGAAAUUGCA_____AAAA ...((((((((....(((((((......))))))).......(..(((....)))..)....))))))))................................................ (-21.46 = -22.34 + 0.88)

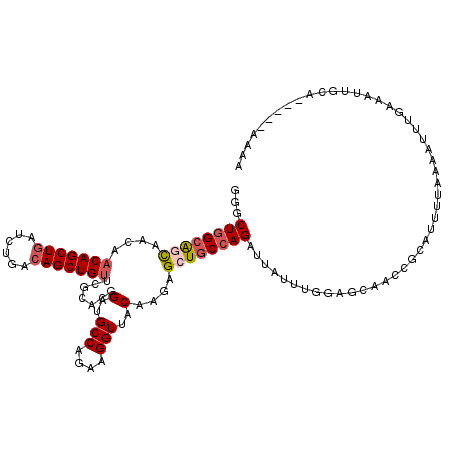
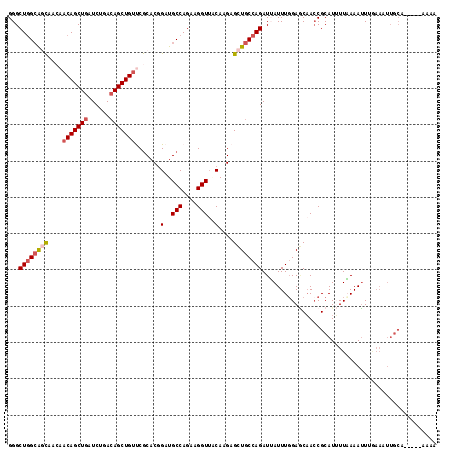
| Location | 7,939,403 – 7,939,515 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -35.86 |
| Consensus MFE | -28.22 |
| Energy contribution | -28.38 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7939403 112 + 20766785 CAAAAUAAUCUGGCAGCUCUUGCAACCUUAUGGCAUCCGUGCGAACAGCUGUCAGAUCAGCUGUUGUUGUUUCCAGCCCUGGUUUGGGAAAAUUUCACGGCCAACAACCGAA .......(((((((((((.(((((.((....))......)))))..)))))))))))..(((((.....((((((((....).)))))))......)))))........... ( -33.60) >DroSec_CAF1 17859 112 + 1 CCAAAUAAUCUGGCAGCUCUUGUAACCUUCUGGCAACGGUGCGGACAGCUUUCAGAUCAGCUGUUGUUGCUGCCAGCCCUGGUUAGGGAAAAUUUCACGGCCAACAACCAAA ((.......((((((((........((..(((....)))...))((((((........))))))....))))))))((((....))))..........))............ ( -33.80) >DroSim_CAF1 19421 112 + 1 CCAAAUAAUCUGGCAGCUCUUGUAACCUUCUGGCAACGGUGCGGACAGCUGUCAGAUCAGCUGUUGUUGCUGCCAGCCCUGGUUAGGGAAAAUUUCACGGCCAACAACCAAA ((.......((((((((........((..(((....)))...))(((((((......)))))))....))))))))((((....))))..........))............ ( -37.50) >DroEre_CAF1 16308 112 + 1 CCAAAUCAUCUGGCAGCUCUUGGAACCUUCUGGCAUCCGUGUGAACAGCUGUCAGAUCAGCUGUUCUUACUGCCAGCCCUGGUUUGGGAAAAUUUCACGGCCAACAACCGAA ((((((((.(((((((.....(((.((....))..)))....(((((((((......)))))))))...)))))))...))))))))..........(((.......))).. ( -42.80) >DroYak_CAF1 23323 112 + 1 CCAAAUUAUCUGGCAUCUCUUGGUACCGUCUGGCAUCCGUGCGAGCAGCUGUCAGAUCAGCUGUUUUUGUCGCUAGCCCUGGUUUGGGAAAAUUUCACGGCCAACAACCGAA ((((((((.(((((........((((.(........).))))(((((((((......))))))))).....)))))...))))))))..........(((.......))).. ( -31.60) >consensus CCAAAUAAUCUGGCAGCUCUUGGAACCUUCUGGCAUCCGUGCGAACAGCUGUCAGAUCAGCUGUUGUUGCUGCCAGCCCUGGUUUGGGAAAAUUUCACGGCCAACAACCGAA .........((((((((...............(((....))).((((((((......))))))))...))))))))(((......)))..........((.......))... (-28.22 = -28.38 + 0.16)

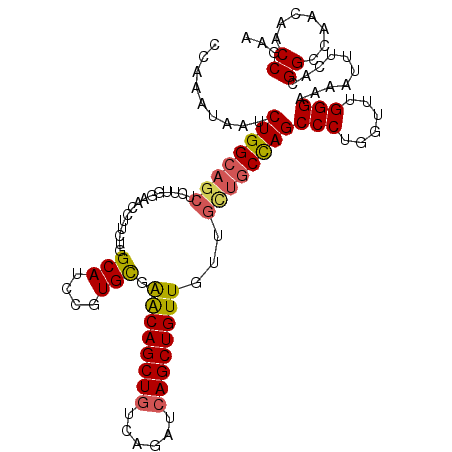
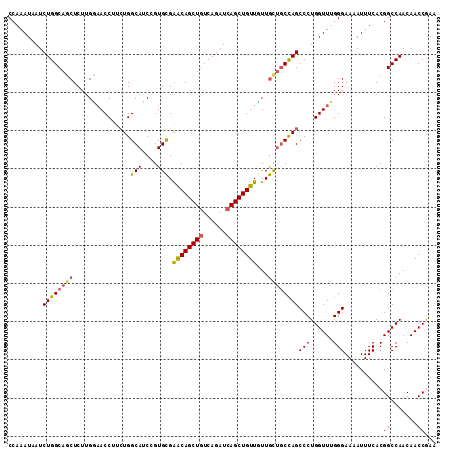
| Location | 7,939,403 – 7,939,515 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -38.85 |
| Consensus MFE | -28.68 |
| Energy contribution | -29.16 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7939403 112 - 20766785 UUCGGUUGUUGGCCGUGAAAUUUUCCCAAACCAGGGCUGGAAACAACAACAGCUGAUCUGACAGCUGUUCGCACGGAUGCCAUAAGGUUGCAAGAGCUGCCAGAUUAUUUUG .(((((((((((((.((..............)).)))((....)).))))))))))((((.(((((.((.(((.....(((....)))))))).))))).))))........ ( -39.84) >DroSec_CAF1 17859 112 - 1 UUUGGUUGUUGGCCGUGAAAUUUUCCCUAACCAGGGCUGGCAGCAACAACAGCUGAUCUGAAAGCUGUCCGCACCGUUGCCAGAAGGUUACAAGAGCUGCCAGAUUAUUUGG ....(((((..(((.((..............)).)))..))))).....(((.(((((((..(((((....)...((.(((....))).))...))))..))))))).))). ( -35.44) >DroSim_CAF1 19421 112 - 1 UUUGGUUGUUGGCCGUGAAAUUUUCCCUAACCAGGGCUGGCAGCAACAACAGCUGAUCUGACAGCUGUCCGCACCGUUGCCAGAAGGUUACAAGAGCUGCCAGAUUAUUUGG ....(((((..(((.((..............)).)))..))))).....(((.(((((((.((((((....)...((.(((....))).))...))))).))))))).))). ( -37.94) >DroEre_CAF1 16308 112 - 1 UUCGGUUGUUGGCCGUGAAAUUUUCCCAAACCAGGGCUGGCAGUAAGAACAGCUGAUCUGACAGCUGUUCACACGGAUGCCAGAAGGUUCCAAGAGCUGCCAGAUGAUUUGG ..((((.....))))..........(((((.((...((((((((..(((((((((......)))))))))....(((.(((....))))))....)))))))).)).))))) ( -45.80) >DroYak_CAF1 23323 112 - 1 UUCGGUUGUUGGCCGUGAAAUUUUCCCAAACCAGGGCUAGCGACAAAAACAGCUGAUCUGACAGCUGCUCGCACGGAUGCCAGACGGUACCAAGAGAUGCCAGAUAAUUUGG ....((((((((((.((..............)).)))))))))).....((((((......))))))(((....((.((((....))))))..)))...(((((...))))) ( -35.24) >consensus UUCGGUUGUUGGCCGUGAAAUUUUCCCAAACCAGGGCUGGCAGCAACAACAGCUGAUCUGACAGCUGUUCGCACGGAUGCCAGAAGGUUACAAGAGCUGCCAGAUUAUUUGG ....((((((((((.((..............)).)))))))))).....(((.(((((((.((((((....)...(..(((....)))..)...))))).))))))).))). (-28.68 = -29.16 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:52 2006