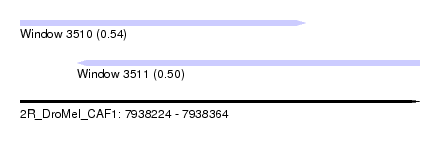
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,938,224 – 7,938,364 |
| Length | 140 |
| Max. P | 0.540339 |

| Location | 7,938,224 – 7,938,324 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -23.11 |
| Energy contribution | -23.28 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
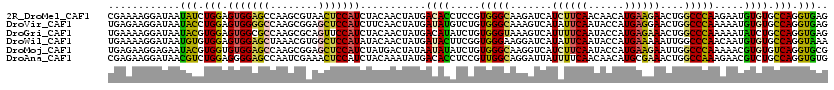
>2R_DroMel_CAF1 7938224 100 + 20766785 CUUAAUAAAGAGAAUCCUU----------GCUCACCUGGCACACAUUCUUGGGCCAGUUCUUCAUGUUGUUGAAGAUGAUCUUGCCCACGGAGGUGUCAUAGUUGUAGAU .........(((.......----------.)))...((((((...(((.(((((.((.((((((......))))))....)).))))).))).))))))........... ( -29.30) >DroSec_CAF1 14966 109 + 1 CUGAAUAAAGAGAAUCCGUUCCGG-GCAGGCUCACCUGGCACACAUUUUUGGGCCAGUUCUUCAUGUUGUUGAAGAUGAUCUUGCCCACGGAGGUGUCAUAGUUGUAGAU .........((.(.(((((...((-(((((.(((.(((((...((....)).))))).((((((......))))))))).))))))))))))..).))............ ( -38.10) >DroSim_CAF1 18237 109 + 1 CUGAAUAAAGAGAAUCCAUUCCGG-GCAGGCUCACCUGGCACACAUUUUUGGGCCAGUUCUUCAUGUUGUUGAAGAUGAUCUUGCCCACGGAGGUGUCAUAGUUGUAGAU ...........((..((..(((((-(((((.(((.(((((...((....)).))))).((((((......))))))))).)))))))..)))))..))............ ( -34.20) >DroEre_CAF1 15134 109 + 1 UCUUAUAUAGGGAAUUAUUUCCGG-ACAGACUCACCUGGCACACAUUCUGGGGCCAGUUCCUCAUGUUAUUGAAGAUGAUCUUGCCCACGGAGGUGUCAUAGUUGUAGAU ..........((((....))))..-.....((((.(((..((((.(((((((((.((.((.(((......))).))....)).)))).)))))))))..))).)).)).. ( -28.40) >DroYak_CAF1 22104 109 + 1 AAAUAUAUAGGGAAUCGUUUCCGC-ACAGACUCACCUGGCACACAUUCUUGGGCCAGUUCUUCAUGUUAUUGAAGAUGAUCUUGCCCACGGAGGUGUCAUAGUUGUAGAU .....(((((((((....))))((-.(((......)))))((((.(((.(((((.((.((((((......))))))....)).))))).))).)))).....)))))... ( -32.50) >DroPer_CAF1 13972 110 + 1 UCUCUUCGGGACGAUCCUUUUCAGCUAACUCUUACCUGGCACACGUUCUUGGGCCAGUUCUUCAUGGUAUUGAAAAUGAUUUUACCUACGGACGUGUCAUAGCUGUAUAU .......(((.....)))...((((((.(........)(.((((((((.((((..((((.((((......))))...))))...)))).))))))))).))))))..... ( -24.50) >consensus CUUAAUAAAGAGAAUCCUUUCCGG_ACAGGCUCACCUGGCACACAUUCUUGGGCCAGUUCUUCAUGUUAUUGAAGAUGAUCUUGCCCACGGAGGUGUCAUAGUUGUAGAU ....................................((((((...(((.(((((.((.((((((......))))))....)).))))).))).))))))........... (-23.11 = -23.28 + 0.17)


| Location | 7,938,244 – 7,938,364 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -22.43 |
| Energy contribution | -21.63 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
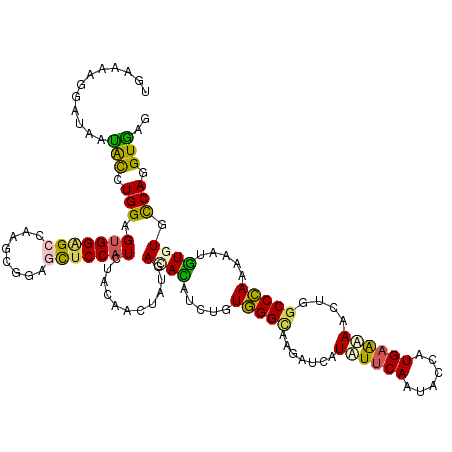
>2R_DroMel_CAF1 7938244 120 - 20766785 CGAAAAGGAUAAUAUCUGGAGUGGAGCCAAGCGUAACUCCAUCUACAACUAUGACACCUCCGUGGGCAAGAUCAUCUUCAACAACAUGAAGAACUGGCCCAAGAAUGUGUGCCAGGUGAG ............(((((((.((((((..........))))))...........((((.....(((((.((....((((((......)))))).)).))))).....)))).))))))).. ( -33.40) >DroVir_CAF1 18455 120 - 1 UGAGAAGGAUAAUACCUGGAGUGGGGCCAAGCGGAGCUCCAUCUUCAACUAUGAUAUGUCUGUGGGCAAAGUCAUAUUCAAUACCAUGAGGAACUGGCCCAAAAAUGUGUGCCAGGUGAG ............(((((((.((.((((((...((....)).((((((..(((((((((.((........)).))))))..)))...))))))..))))))....)).....))))))).. ( -35.30) >DroGri_CAF1 15980 120 - 1 UGAAAAGGAUAAUACGUGGAGUGGCGCCAAGCGCAGUUCCAUCUACAACUAUGACAUAUCUGUGGGUAAAGUCAUUUUCAAUACCAUGAGAAACUGGCCCAAAAAUAUCUGCCAGGUGAG ((((((.(((..(((((((((..((((...))))..))))))(((((..(((...)))..))))))))..))).)))))).............(((((............)))))..... ( -30.30) >DroWil_CAF1 18153 120 - 1 UGAAAAGGAUAAUGUGUGGAGUGGAGCUAAACGUGGCUCCAUAUACAACUAUGAUACUUCGGUGGGAAGGAUCAUAUUCAAUACCAUGAAAAAUUGGCCCAACAAUGUGUGCCAGGUAAA ((((..........((((..(((((((((....))))))))).))))..((((((.((((.....)))).)))))))))).((((.(....)..((((.((......)).)))))))).. ( -34.40) >DroMoj_CAF1 20348 120 - 1 UGAGAAGGAGAAUACGUGGUGUGGAGCCAAGCGGAGCUCCAUCUAUGACUAUAAUAUAUCUGUGGGCAAGGUCAUCUUCAAUACCAUGAAGAAUUGGCCCAAAAACGUGUGUCAGGUGCG ..............(((((.(((((((........))))))))))))(((...((((((.(.(((((.......((((((......))))))....)))))...).))))))..)))... ( -32.30) >DroAna_CAF1 19838 120 - 1 CGAGAAGGAUAACGUCUGGAGGGGAGCCAAUCGAAACUCCAUCUACAAAUAUGACACCUCCGUUGGCAGGAUUAUUUUCAACAACAUGCGAAACUGGCCAAAGAACGUCUGCCAGGUGUG .(((((.(((...(((((((((((((..........)))).....((....))...))))))..)))...))).)))))........(((...(((((...((.....))))))).))). ( -28.10) >consensus UGAAAAGGAUAAUACCUGGAGUGGAGCCAAGCGGAGCUCCAUCUACAACUAUGACACAUCUGUGGGCAAGAUCAUAUUCAAUACCAUGAAAAACUGGCCCAAAAAUGUGUGCCAGGUGAG ............(((.(((.(((((((........)))))))...........((((.....(((((.......((((((......))))))....))))).....)))).))).))).. (-22.43 = -21.63 + -0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:48 2006