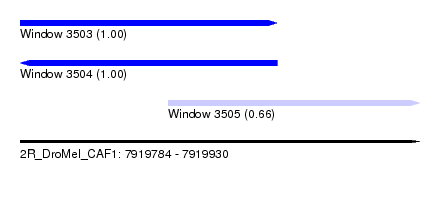
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,919,784 – 7,919,930 |
| Length | 146 |
| Max. P | 0.999779 |

| Location | 7,919,784 – 7,919,878 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 93.01 |
| Mean single sequence MFE | -16.03 |
| Consensus MFE | -14.04 |
| Energy contribution | -14.28 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.997096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
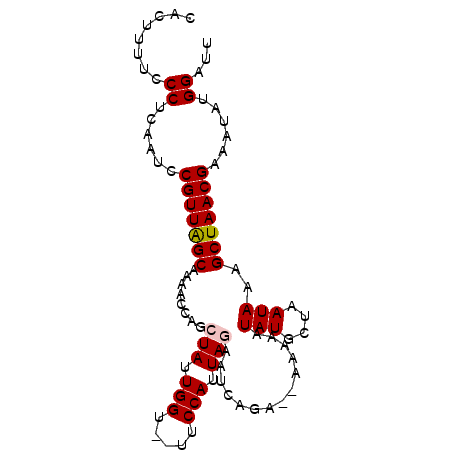
>2R_DroMel_CAF1 7919784 94 + 20766785 CACUUUUCCCUCAAUCCGUUAGCAAAACCAGCUAUUGGUGCUUCCAUUAGAAUAAAAAAAAAAAUAUGCUAAUAAAGCUAACGAAAUAUGGAUU ......(((.......(((((((........(((.(((.....))).)))..............(((....)))..)))))))......))).. ( -15.92) >DroSec_CAF1 16787 90 + 1 CACUUUUCCCUCAAUCCGUUAGCAAAACCAGCUAUUGGU--UUCCAUUAGAAUCAGA--AAAAAUAUGCUAAUAAAGCUAACGAAAUAUGGAUU ......(((.......(((((((.(((((((...)))))--))..(((((.......--.........)))))...)))))))......))).. ( -16.21) >DroSim_CAF1 17815 90 + 1 CACUUUUCCCUCAAUCCGUUGGCAAAACCAGCUAUUGGU--UUCCAUUAGAAUCAGA--AAAAAUAUGCUAAUAAAGCUAACGAAAUAUGGAUU ......(((.......(((((((.(((((((...)))))--))..(((((.......--.........)))))...)))))))......))).. ( -15.91) >DroEre_CAF1 16778 89 + 1 CACUU-UCCCUUAACCCGUUAGCAAAACCAGCUAUUGGU--UUCCAUUAAAAUCAGA--AAAAAUAUGCUAAUAAAGCUAACGAAUUAUGGAUU .....-(((..(((..(((((((......(((((((..(--(((...........))--)).)))).)))......)))))))..))).))).. ( -17.20) >DroYak_CAF1 19726 89 + 1 CACUU-UCCCUUAAUCCGUUAGCAAAACCAGCUAUUGGU--UUCCAUUAAAAUCAAA--CAGAAUAUGCUAAUAAAGCUAACGAAAUAUGGAUU .....-(((.......(((((((.(((((((...)))))--)).........((...--..)).............)))))))......))).. ( -14.92) >consensus CACUUUUCCCUCAAUCCGUUAGCAAAACCAGCUAUUGGU__UUCCAUUAGAAUCAGA__AAAAAUAUGCUAAUAAAGCUAACGAAAUAUGGAUU ........((......(((((((........(((.(((.....))).)))..............(((....)))..)))))))......))... (-14.04 = -14.28 + 0.24)

| Location | 7,919,784 – 7,919,878 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 93.01 |
| Mean single sequence MFE | -24.56 |
| Consensus MFE | -21.32 |
| Energy contribution | -20.88 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7919784 94 - 20766785 AAUCCAUAUUUCGUUAGCUUUAUUAGCAUAUUUUUUUUUUUAUUCUAAUGGAAGCACCAAUAGCUGGUUUUGCUAACGGAUUGAGGGAAAAGUG ..(((....(((((((((...((((((.((((....(((((((....)))))))....))))))))))...))))))))).....)))...... ( -24.70) >DroSec_CAF1 16787 90 - 1 AAUCCAUAUUUCGUUAGCUUUAUUAGCAUAUUUUU--UCUGAUUCUAAUGGAA--ACCAAUAGCUGGUUUUGCUAACGGAUUGAGGGAAAAGUG ..(((....(((((((((...((((((.((((.((--(((.........))))--)..))))))))))...))))))))).....)))...... ( -25.20) >DroSim_CAF1 17815 90 - 1 AAUCCAUAUUUCGUUAGCUUUAUUAGCAUAUUUUU--UCUGAUUCUAAUGGAA--ACCAAUAGCUGGUUUUGCCAACGGAUUGAGGGAAAAGUG ..(((....((((((.((...((((((.((((.((--(((.........))))--)..))))))))))...)).)))))).....)))...... ( -20.70) >DroEre_CAF1 16778 89 - 1 AAUCCAUAAUUCGUUAGCUUUAUUAGCAUAUUUUU--UCUGAUUUUAAUGGAA--ACCAAUAGCUGGUUUUGCUAACGGGUUAAGGGA-AAGUG ..(((.((((((((((((...((((((.((((.((--(((.........))))--)..))))))))))...)))))))))))).))).-..... ( -27.80) >DroYak_CAF1 19726 89 - 1 AAUCCAUAUUUCGUUAGCUUUAUUAGCAUAUUCUG--UUUGAUUUUAAUGGAA--ACCAAUAGCUGGUUUUGCUAACGGAUUAAGGGA-AAGUG ..(((....(((((((((...((((((.((((...--(((.((....)).)))--...))))))))))...))))))))).....)))-..... ( -24.40) >consensus AAUCCAUAUUUCGUUAGCUUUAUUAGCAUAUUUUU__UCUGAUUCUAAUGGAA__ACCAAUAGCUGGUUUUGCUAACGGAUUGAGGGAAAAGUG ..(((.((.(((((((((...((((((.((((.....(((.........)))......))))))))))...))))))))).)).)))....... (-21.32 = -20.88 + -0.44)


| Location | 7,919,838 – 7,919,930 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.20 |
| Mean single sequence MFE | -14.39 |
| Consensus MFE | -9.64 |
| Energy contribution | -10.83 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7919838 92 + 20766785 AAAAAAAAAAUAUGCUAAUAAAGCUAACGAAAUAUGGAUUAGUGAGCCAUAUUUAAAUAGA---------------------GGUCUGCGCAGAAAUGCUAAAAGUUAAACAG---- .............((..((....(((...((((((((.........))))))))...))).---------------------.))..))(((....)))..............---- ( -15.50) >DroSec_CAF1 16839 90 + 1 AGA--AAAAAUAUGCUAAUAAAGCUAACGAAAUAUGGAUUAGUGAGCCAUAUUUAAAUAGA---------------------AGUCUGCGCAGAAAUGCUAAAAGUUAAACAG---- ...--........((..((....(((...((((((((.........))))))))...))).---------------------.))..))(((....)))..............---- ( -14.40) >DroSim_CAF1 17867 90 + 1 AGA--AAAAAUAUGCUAAUAAAGCUAACGAAAUAUGGAUUAGUGAGCCAUAUUUAAAUAGA---------------------AGUCUGCGCAGAAAUGCUAAAAGUUAAACAG---- ...--........((..((....(((...((((((((.........))))))))...))).---------------------.))..))(((....)))..............---- ( -14.40) >DroEre_CAF1 16829 90 + 1 AGA--AAAAAUAUGCUAAUAAAGCUAACGAAUUAUGGAUUAGUGAGCCAUAUUUAAAUAGA---------------------GGUCUGCGCAGAAAUGCUAAAAGUUGAACAA---- ...--........(((.....)))((((.............((..(((.(((....)))..---------------------)))..))(((....))).....)))).....---- ( -12.60) >DroYak_CAF1 19777 90 + 1 AAA--CAGAAUAUGCUAAUAAAGCUAACGAAAUAUGGAUUAGUGAGCCAUAUUUAAAUAGA---------------------AGUCUGCGCAGAAAUGCUAAAAGUUAAACAA---- ...--((((....(((.....))).....((((((((.........)))))))).......---------------------..)))).(((....)))..............---- ( -15.70) >DroAna_CAF1 16020 113 + 1 --G--CAGCUUAUGCUAAUAGAGAUAAUGAAAAGUAGAUUAAUUAACCAUUUUUAAAUGAAAGUCAGCUCUUAAUGAAAAGUAGUCUGCACACAAAUGUUAAAUGUUAAAUAAAGAG --(--((.....)))(((((..((((.......((((((((......((((......((.....))......)))).....)))))))).......))))...)))))......... ( -13.74) >consensus AGA__AAAAAUAUGCUAAUAAAGCUAACGAAAUAUGGAUUAGUGAGCCAUAUUUAAAUAGA_____________________AGUCUGCGCAGAAAUGCUAAAAGUUAAACAA____ .............(((.....)))((((.((((((((.........))))))))...................................(((....))).....))))......... ( -9.64 = -10.83 + 1.20)

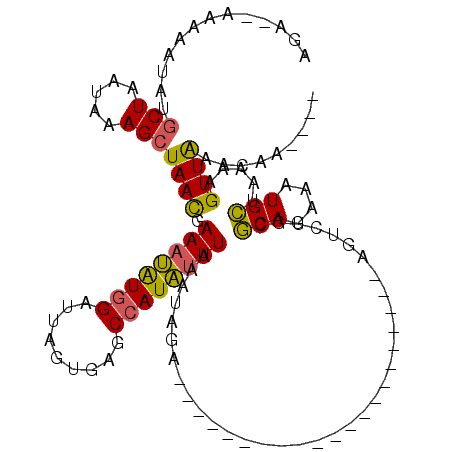
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:42 2006