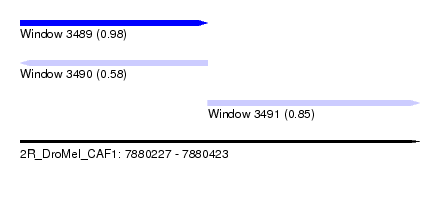
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,880,227 – 7,880,423 |
| Length | 196 |
| Max. P | 0.981390 |

| Location | 7,880,227 – 7,880,319 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 73.76 |
| Mean single sequence MFE | -15.90 |
| Consensus MFE | -8.60 |
| Energy contribution | -9.27 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
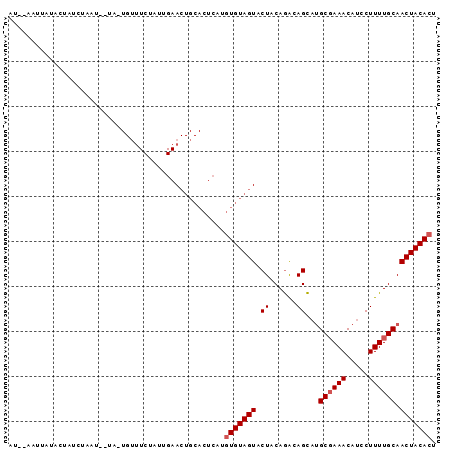
>2R_DroMel_CAF1 7880227 92 + 20766785 AU--AAUUAUAGUAUCUAAUUUUA-UGUUUCUAUUGAACUGCACUCAUGUGUAGUACUACAGACAGCAUGCGAAACAUCCUUUUGCAACUACACU ..--.....((((..........(-(((((((...(.(((((((....))))))).)...))).)))))((((((.....)))))).)))).... ( -19.70) >DroSec_CAF1 12210 93 + 1 AU--AAUUAUACUAUCUAAUAGUAUUGUUUCUAUUGAACUGCACUCAUGUGUAGUACUACAGGCAGCAUGCUAAACAUCCUUUUGCAACUACACU ..--...............((((((.((((((...(.(((((((....))))))).)...))).)))))))))...................... ( -16.50) >DroEre_CAF1 9724 73 + 1 AUUGAAAUAAACUA-------------CAUCUAUUGAA---------UCUGUAGUACUACAAACAGUUCGCGAAACAUCCUUUUGCAACUACACU ..........((((-------------((((....)).---------..)))))).........((((.((((((.....))))))))))..... ( -11.50) >consensus AU__AAUUAUACUAUCUAAU__UA_UGUUUCUAUUGAACUGCACUCAUGUGUAGUACUACAGACAGCAUGCGAAACAUCCUUUUGCAACUACACU ................................................(((((((.((......))...((((((.....)))))).))))))). ( -8.60 = -9.27 + 0.67)

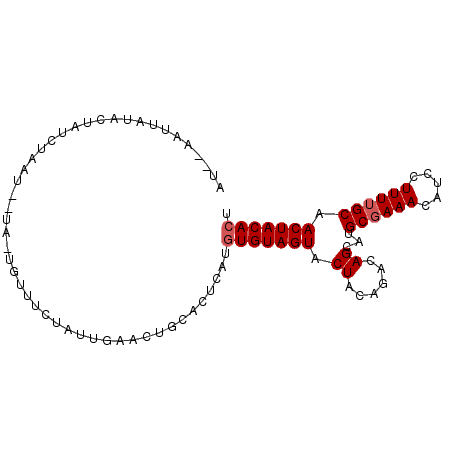
| Location | 7,880,227 – 7,880,319 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 73.76 |
| Mean single sequence MFE | -17.14 |
| Consensus MFE | -12.82 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7880227 92 - 20766785 AGUGUAGUUGCAAAAGGAUGUUUCGCAUGCUGUCUGUAGUACUACACAUGAGUGCAGUUCAAUAGAAACA-UAAAAUUAGAUACUAUAAUU--AU ..((((((.........(((((((...(((.....)))(((((.(....)))))).........))))))-)..........))))))...--.. ( -17.11) >DroSec_CAF1 12210 93 - 1 AGUGUAGUUGCAAAAGGAUGUUUAGCAUGCUGCCUGUAGUACUACACAUGAGUGCAGUUCAAUAGAAACAAUACUAUUAGAUAGUAUAAUU--AU .(((((((((((..((.(((.....))).))...))))..)))))))....((((.((..(((((........)))))..)).))))....--.. ( -22.70) >DroEre_CAF1 9724 73 - 1 AGUGUAGUUGCAAAAGGAUGUUUCGCGAACUGUUUGUAGUACUACAGA---------UUCAAUAGAUG-------------UAGUUUAUUUCAAU ..((((((((((((((..((.....))..)).))))))..))))))((---------...(((((((.-------------..)))))))))... ( -11.60) >consensus AGUGUAGUUGCAAAAGGAUGUUUCGCAUGCUGUCUGUAGUACUACACAUGAGUGCAGUUCAAUAGAAACA_UA__AUUAGAUAGUAUAAUU__AU .(((((((((((..((.(((.....))).))...))))..)))))))................................................ (-12.82 = -13.27 + 0.45)

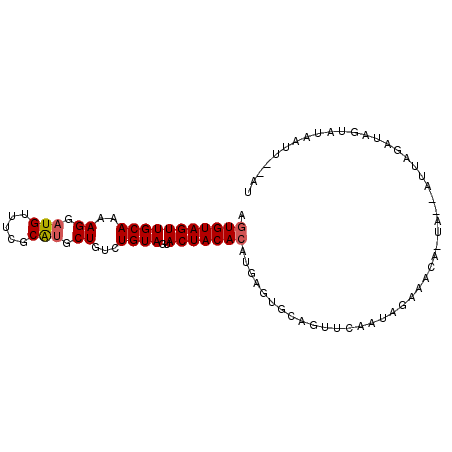
| Location | 7,880,319 – 7,880,423 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 72.79 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -17.55 |
| Energy contribution | -15.92 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7880319 104 + 20766785 AGCCGAGGAAACGUGGCAGGCUGGAAUUAUACCAUCCGACACAGAUUUCCGAAUUUUUCGGGACUUCGGAAACGUGCCCGGUAAGCUUAUAUGGUUAUGAAC-CA .((((.(....).))))(((((((.......))..(((.(((.((.((((((.....))))))..))(....))))..)))..)))))...((((.....))-)) ( -30.90) >DroPse_CAF1 8643 97 + 1 GGCUGAGGAGAUCUUUCAGAGUGGAAUUUUGCCAUCUGAUUCAGAUUUCCGAAUUUUCCGUGACUAUGGAGAUGUUCCCGGUGAGUAACUA-------AAAC-AC .((((.((((((((.(((((.(((.......))))))))...))))))))(((((((((((....))))))).)))).)))).........-------....-.. ( -29.80) >DroSec_CAF1 12303 104 + 1 AGCCGAGGAAACGUGGCAGGCUGGUAUUAUACCAUCCGACACGGAUUUCCGCAUUUUUCGGGACUUCGGAAACGUGCCUGGUAAGCUUAUAUUGCCAUGAAC-UA ..((((((...((((...((.(((((...))))).))..))))...(((((.......)))))))))))....((.(.((((((.......)))))).).))-.. ( -32.20) >DroEre_CAF1 9797 104 + 1 AGCUGAGGAGACCUGGCAGGCUGGAAUUAUACCAUCUGACACGGAUUUCCGUAUCUUUCGGGACUUCGGAAACGUGCCAGGUCAGCUUCUACUGCAAUAAAC-UG .((.((((.(((((((((.............((....((.((((....)))).)).....))....((....)))))))))))..))))....)).......-.. ( -35.00) >DroAna_CAF1 9640 103 + 1 GGCCGAAGAGAUUUUUCAGAGUGGAAUUCUACCCUCUGACUCAGAUUUUCGUAUUUUUCGAGACUUUGGAAAUAUACCCGGUCAGU--AAUGAGUCAUAAACAUU (((((..((((.(((((((((.((.......))))))))...))).))))(((((((((((....))))))).)))).)))))...--................. ( -24.30) >DroPer_CAF1 9599 97 + 1 GGCUGAGGAGAUCUUUCAGAGUGGAAUUUUGCCAUCUGAUUCAGAUUUCCGAAUUUUCCGUGACUAUGGAGAUGUUCCUGGUGAGUAACUA-------AAAC-AC .(((.(((((((((.(((((.(((.......))))))))...))))))))(((((((((((....))))))).))))....).))).....-------....-.. ( -27.90) >consensus AGCCGAGGAGACCUGGCAGACUGGAAUUAUACCAUCUGACACAGAUUUCCGAAUUUUUCGGGACUUCGGAAACGUGCCCGGUAAGCUUAUA__G__AUAAAC_UA .((((.(((((.((..((((.(((.......)))))))....)).)))))(((((((((((....))))))).)))).))))....................... (-17.55 = -15.92 + -1.63)

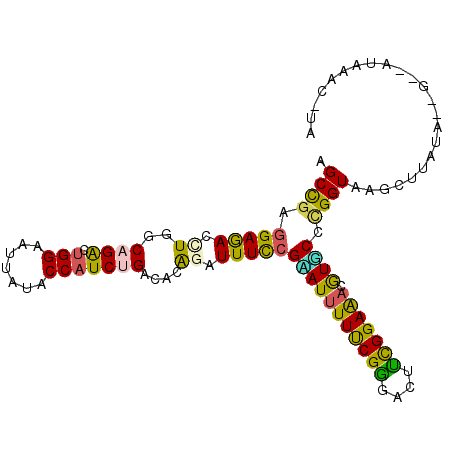
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:27 2006