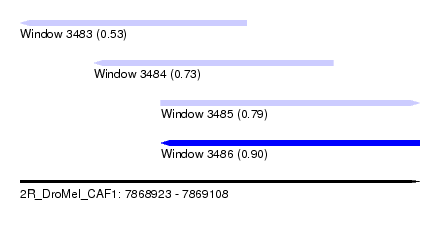
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,868,923 – 7,869,108 |
| Length | 185 |
| Max. P | 0.904562 |

| Location | 7,868,923 – 7,869,028 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.38 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7868923 105 - 20766785 UCGGAUGGCAAAAUGCCAGACUGAAAUAUUUCCUCGGCCAUUGUGGUGGCAAACGGAUGUUUAGCAUUCUGGUAAUAGUACUAUGGGUAGUAAAUGAAGUAAAUA ((((.((((.....))))..))))..((((((....(((((....)))))....(((((.....))))).........(((((....)))))...)))))).... ( -25.80) >DroSim_CAF1 11589 105 - 1 UCGGAUGGCAAAAUGCCAGACUGAAAUAUUUCCUCGGCCAUUGUGGUGGCAAAUGGAUGUUUAGCAUUCUGGUAAUAGUACUACGGGUAGUAAAUGAAAUAAAUA ((((.((((.....))))..))))..(((((((((((((((....)))))....(((((.....)))))(((((....)))))))))........)))))).... ( -26.80) >DroEre_CAF1 12983 105 - 1 UCGGAUGGUAAAAUGCCAGACUGAAAUAUUUCCUCGGCCAUUGUGGUGGCAAAUGGGUGUUUCGCAUUCUGGUAAUAGUACUAUGGGGAGUAAAUAAAAUAAAUG ((..((((((...(((((((.((((((((..((...(((((....)))))....))))))))))...)))))))....))))))..))................. ( -28.40) >DroYak_CAF1 11868 105 - 1 UCUGAUGGUAAAAUGCCAGACUGAAAUAUUUCCUCGGCCAUUGUGGUGGCAAACGGAUGUUUCGCAUUCUGGUAAUAGUACUAUGGGGAGUAAAUAAAAUAAAUA (((.((((((...(((((((.((((((((((.....(((((....)))))....))))))))))...)))))))....)))))).)))................. ( -31.90) >DroAna_CAF1 11226 98 - 1 UCAGAGGGCAGGAUUCCUGAUUGAAAAAUCUCUUCGGCCAUUGUGGUGCCAAACGGAUGUUUAGCAUUCUGGUUGUAAUACUGUAA-------AAUAAUUGUAAA ((((((.((..((((((.((((....)))).....((((......).)))....))).)))..)).))))))..............-------............ ( -20.10) >consensus UCGGAUGGCAAAAUGCCAGACUGAAAUAUUUCCUCGGCCAUUGUGGUGGCAAACGGAUGUUUAGCAUUCUGGUAAUAGUACUAUGGGGAGUAAAUAAAAUAAAUA .............(((((((.((....((.(((...(((((....)))))....))).))....)).)))))))............................... (-18.30 = -18.78 + 0.48)

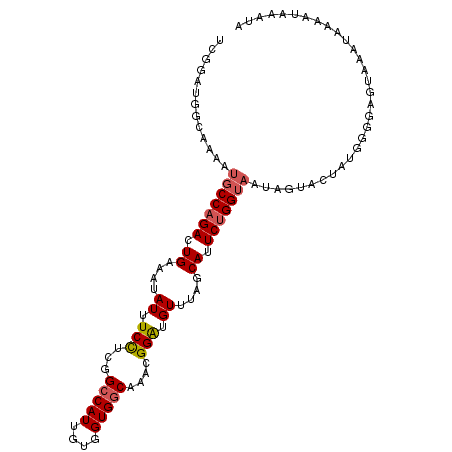
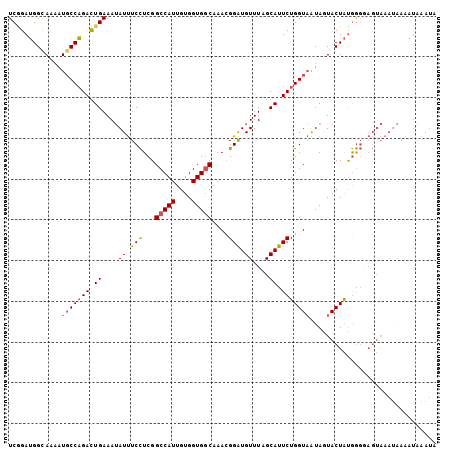
| Location | 7,868,957 – 7,869,068 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -35.26 |
| Consensus MFE | -27.14 |
| Energy contribution | -27.34 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7868957 111 - 20766785 UGGCAGUUGUCCGUAGUCGCGAAAGAUACGGAAAUCCGUAUCGGAUGGCAAAAUGCCAGACUGAAAUAUUUCCUCGGCCAUUGUGGUGGCAAACGGAUGUUUAGCAUUCUG (((((((..(((((....))....(((((((....))))))))))..))....)))))(.(((((.....(((...(((((....)))))....)))..))))))...... ( -38.30) >DroSim_CAF1 11623 111 - 1 CGGCAGUUGUCCGUAGUCGCGAAAGAUACGGAAAUCCGUAUCGGAUGGCAAAAUGCCAGACUGAAAUAUUUCCUCGGCCAUUGUGGUGGCAAAUGGAUGUUUAGCAUUCUG .((((((..(((((....))....(((((((....))))))))))..))....))))(((.((....((.(((...(((((....)))))....))).))....)).))). ( -39.00) >DroEre_CAF1 13017 111 - 1 UGGCAGUUGUCCGUAGUCGCGAAAGAUACGGAAAUCCGUAUCGGAUGGUAAAAUGCCAGACUGAAAUAUUUCCUCGGCCAUUGUGGUGGCAAAUGGGUGUUUCGCAUUCUG (((((.(..(((((....))....(((((((....))))))))))..).....)))))....(((((((..((...(((((....)))))....)))))))))........ ( -36.80) >DroYak_CAF1 11902 111 - 1 UGGCAGUUGUCCGUAGUCGCGAAAGAUACGGAAAUCCGUAUCUGAUGGUAAAAUGCCAGACUGAAAUAUUUCCUCGGCCAUUGUGGUGGCAAACGGAUGUUUCGCAUUCUG (((((.....((((.........((((((((....)))))))).)))).....)))))....(((((((((.....(((((....)))))....)))))))))........ ( -36.40) >DroAna_CAF1 11253 111 - 1 CGGAAGCUGGCCGUAAUCCCGAAAUAUACGAAAAUCAGAAUCAGAGGGCAGGAUUCCUGAUUGAAAAAUCUCUUCGGCCAUUGUGGUGCCAAACGGAUGUUUAGCAUUCUG (((((((((((((((((.(((((.........((((((((((.........)))).))))))..........)))))..)))))))).((....)).....)))).))))) ( -25.81) >consensus UGGCAGUUGUCCGUAGUCGCGAAAGAUACGGAAAUCCGUAUCGGAUGGCAAAAUGCCAGACUGAAAUAUUUCCUCGGCCAUUGUGGUGGCAAACGGAUGUUUAGCAUUCUG .((((.(((.((((..........(((((((....)))))))..)))))))..))))(((.((....((.(((...(((((....)))))....))).))....)).))). (-27.14 = -27.34 + 0.20)

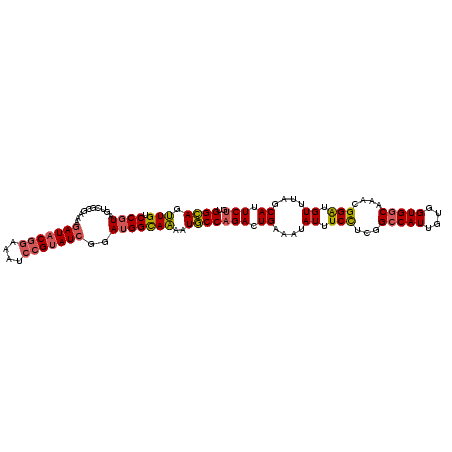
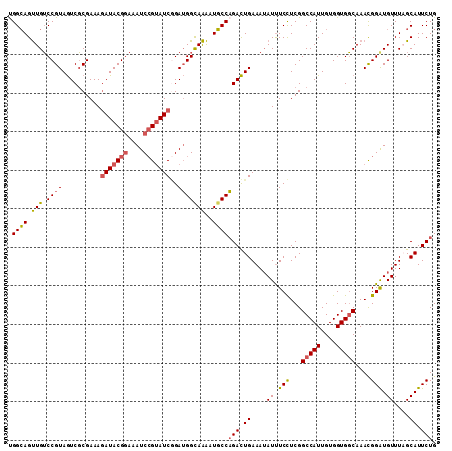
| Location | 7,868,988 – 7,869,108 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -40.12 |
| Consensus MFE | -31.29 |
| Energy contribution | -31.10 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7868988 120 + 20766785 UGGCCGAGGAAAUAUUUCAGUCUGGCAUUUUGCCAUCCGAUACGGAUUUCCGUAUCUUUCGCGACUACGGACAACUGCCAGGUGAGUUGGUCACGCGAAUGCUUCAUUGGCUUGUGUAUU .((((((.(((........(..((((.....))))..)(((((((....))))))).((((((.(....).(((((........)))))....))))))...))).))))))........ ( -39.20) >DroSim_CAF1 11654 120 + 1 UGGCCGAGGAAAUAUUUCAGUCUGGCAUUUUGCCAUCCGAUACGGAUUUCCGUAUCUUUCGCGACUACGGACAACUGCCGGGUGAGUUGGUCACGCGAAUGCUUCAUUCGCUUGUGUACU (((((((........((((.(((((((..((((.....(((((((....)))))))....))))(....).....)))))))))))))))))).(((((((...)))))))......... ( -39.50) >DroEre_CAF1 13048 119 + 1 UGGCCGAGGAAAUAUUUCAGUCUGGCAUUUUACCAUCCGAUACGGAUUUCCGUAUCUUUCGCGACUACGGACAACUGCCAGGUGAGUUGGUCACGCGAGUGCCUUACUGGUUUGGU-AUU ..((((((........(((((..(((((..........(((((((....)))))))..(((((.(....).(((((........)))))....))))))))))..)))))))))))-... ( -39.80) >DroYak_CAF1 11933 120 + 1 UGGCCGAGGAAAUAUUUCAGUCUGGCAUUUUACCAUCAGAUACGGAUUUCCGUAUCUUUCGCGACUACGGACAACUGCCAGGUGAGUUGGUCACGCGAAUGCCUUUCUGCUUUCCUCAUU .....(((((((.....(((...((((..........((((((((....))))))))((((((.(....).(((((........)))))....))))))))))...))).)))))))... ( -42.00) >consensus UGGCCGAGGAAAUAUUUCAGUCUGGCAUUUUACCAUCCGAUACGGAUUUCCGUAUCUUUCGCGACUACGGACAACUGCCAGGUGAGUUGGUCACGCGAAUGCCUCACUGGCUUGUGUAUU ..(((((.(((....)))..)).))).....((((...(((((((....))))))).((((((...((.(((.(((....)))..))).))..))))))........))))......... (-31.29 = -31.10 + -0.19)

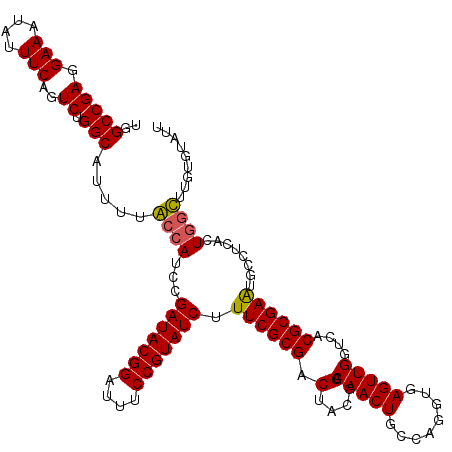
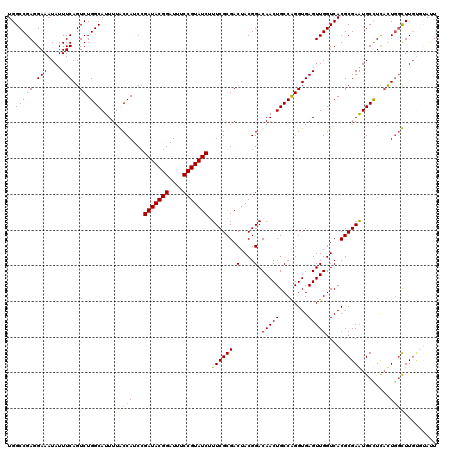
| Location | 7,868,988 – 7,869,108 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -29.98 |
| Energy contribution | -30.22 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7868988 120 - 20766785 AAUACACAAGCCAAUGAAGCAUUCGCGUGACCAACUCACCUGGCAGUUGUCCGUAGUCGCGAAAGAUACGGAAAUCCGUAUCGGAUGGCAAAAUGCCAGACUGAAAUAUUUCCUCGGCCA .........(((...((((..(((..((((.....))))((((((((..(((((....))....(((((((....))))))))))..))....))))))...)))...))))...))).. ( -37.40) >DroSim_CAF1 11654 120 - 1 AGUACACAAGCGAAUGAAGCAUUCGCGUGACCAACUCACCCGGCAGUUGUCCGUAGUCGCGAAAGAUACGGAAAUCCGUAUCGGAUGGCAAAAUGCCAGACUGAAAUAUUUCCUCGGCCA ..(((....(((((((...)))))))(.(((.((((........)))))))))))((((.(((((((((((....)))))))(..((((.....))))..).......))))..)))).. ( -38.10) >DroEre_CAF1 13048 119 - 1 AAU-ACCAAACCAGUAAGGCACUCGCGUGACCAACUCACCUGGCAGUUGUCCGUAGUCGCGAAAGAUACGGAAAUCCGUAUCGGAUGGUAAAAUGCCAGACUGAAAUAUUUCCUCGGCCA ...-.......((((..(((((....))).)).......((((((.(..(((((....))....(((((((....))))))))))..).....))))))))))................. ( -34.70) >DroYak_CAF1 11933 120 - 1 AAUGAGGAAAGCAGAAAGGCAUUCGCGUGACCAACUCACCUGGCAGUUGUCCGUAGUCGCGAAAGAUACGGAAAUCCGUAUCUGAUGGUAAAAUGCCAGACUGAAAUAUUUCCUCGGCCA ..((((((((.(((...((((((((((..(((((((........)))))...))...))))))((((((((....))))))))..........))))...))).....)))))))).... ( -41.80) >consensus AAUAAACAAACCAAUAAAGCAUUCGCGUGACCAACUCACCUGGCAGUUGUCCGUAGUCGCGAAAGAUACGGAAAUCCGUAUCGGAUGGCAAAAUGCCAGACUGAAAUAUUUCCUCGGCCA .................(((....(((.(((.((((........))))))))))((((......(((((((....)))))))...((((.....))))))))..............))). (-29.98 = -30.22 + 0.25)

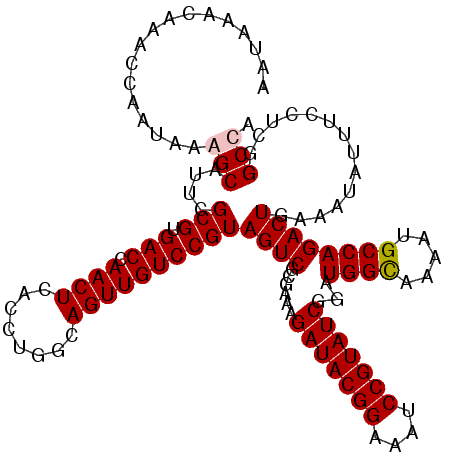
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:22 2006