
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
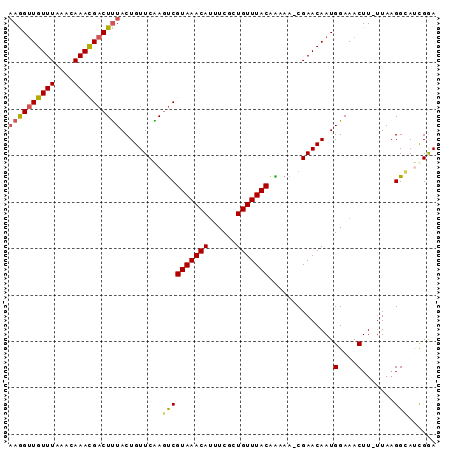
| Location | 7,858,491 – 7,858,581 |
| Length | 90 |
| Max. P | 0.990423 |

| Location | 7,858,491 – 7,858,581 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 88.36 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -15.42 |
| Energy contribution | -15.54 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7858491 90 + 20766785 AAGGUUGUUUAAACAAAUGACUUUACUGUUCAACUCGUAAACAUUUCACUGUUUACAAAAA-CGAACAAUGGAAACUU-GUAUGGAAUUGGA (((((..(((....)))..)))))....(((((.(((((((((......))))))).....-((.((((.(....)))-)).)))).))))) ( -19.60) >DroSec_CAF1 1383 91 + 1 AAAGUUGUUUAAACAAACGACUUUACUGUUCAAGUCGUAAACAUUUCGCUGUUUACAAAAAGCGAACAAUGGAAACUU-UUAAGGCAUCGGA ((((((((((....)))))))))).(((.....(((.((((...((((((..........))))))....(....).)-))).)))..))). ( -18.70) >DroSim_CAF1 1375 91 + 1 UAAGUUGUUUAAACAAACGACUUUACUGUUCAAGUCGUAAACAUUUCGCUGUUUACAAAAAGCGAACAAUGGAAACUU-UUAAGGCAUCGGA .(((((((((....)))))))))..(((.....(((.((((...((((((..........))))))....(....).)-))).)))..))). ( -17.40) >DroEre_CAF1 2567 91 + 1 AAGGCUGUUUAAACAAACGACUUUACUGUUCGAGUCGUAAACAUUUCGCUGUUUACAGCAA-CGAACAAUGGAAACUUUUUAAGGCAACGAA ...(((((((......(((((((........))))))).........((((....))))..-.))))...(....).......)))...... ( -19.60) >DroYak_CAF1 1394 90 + 1 AAGGUUGUUUAAAUAAACGACUCUACUGUUCGAGCCGUAAACAAUUCGCUGUUUACGGCAA-CGAACAAUGGAAACUU-UUAAGGUACCGAA ...(((((((....)))))))((((.((((((.((((((((((......))))))))))..-)))))).)))).....-............. ( -30.50) >consensus AAGGUUGUUUAAACAAACGACUUUACUGUUCAAGUCGUAAACAUUUCGCUGUUUACAAAAA_CGAACAAUGGAAACUU_UUAAGGCAUCGGA ((((((((((....)))))))))).........((((((((((......)))))))..............(....).......)))...... (-15.42 = -15.54 + 0.12)


| Location | 7,858,491 – 7,858,581 |
|---|---|
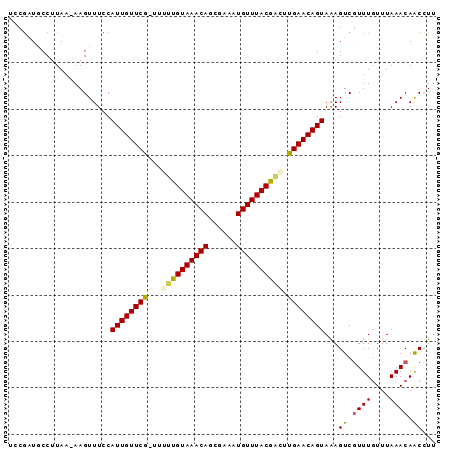
| Length | 90 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 88.36 |
| Mean single sequence MFE | -21.34 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.42 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7858491 90 - 20766785 UCCAAUUCCAUAC-AAGUUUCCAUUGUUCG-UUUUUGUAAACAGUGAAAUGUUUACGAGUUGAACAGUAAAGUCAUUUGUUUAAACAACCUU ...........((-((((..(.((((((((-..((((((((((......)))))))))).))))))))...)..))))))............ ( -19.40) >DroSec_CAF1 1383 91 - 1 UCCGAUGCCUUAA-AAGUUUCCAUUGUUCGCUUUUUGUAAACAGCGAAAUGUUUACGACUUGAACAGUAAAGUCGUUUGUUUAAACAACUUU .............-........((((((((....(((((((((......)))))))))..))))))))(((((.((((....)))).))))) ( -18.40) >DroSim_CAF1 1375 91 - 1 UCCGAUGCCUUAA-AAGUUUCCAUUGUUCGCUUUUUGUAAACAGCGAAAUGUUUACGACUUGAACAGUAAAGUCGUUUGUUUAAACAACUUA .............-........((((((((....(((((((((......)))))))))..)))))))).((((.((((....)))).)))). ( -18.30) >DroEre_CAF1 2567 91 - 1 UUCGUUGCCUUAAAAAGUUUCCAUUGUUCG-UUGCUGUAAACAGCGAAAUGUUUACGACUCGAACAGUAAAGUCGUUUGUUUAAACAGCCUU ...((((..(((((..(.....((((((((-..(.((((((((......)))))))).).)))))))).....).....))))).))))... ( -19.50) >DroYak_CAF1 1394 90 - 1 UUCGGUACCUUAA-AAGUUUCCAUUGUUCG-UUGCCGUAAACAGCGAAUUGUUUACGGCUCGAACAGUAGAGUCGUUUAUUUAAACAACCUU ...(((.......-..(.(((.((((((((-..(((((((((((....))))))))))).)))))))).))).)((((....)))).))).. ( -31.10) >consensus UCCGAUGCCUUAA_AAGUUUCCAUUGUUCG_UUUUUGUAAACAGCGAAAUGUUUACGACUUGAACAGUAAAGUCGUUUGUUUAAACAACCUU ......................((((((((...((((((((((......)))))))))).))))))))...((.((((....)))).))... (-18.58 = -18.42 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:15 2006