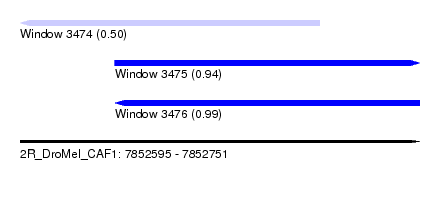
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,852,595 – 7,852,751 |
| Length | 156 |
| Max. P | 0.993505 |

| Location | 7,852,595 – 7,852,712 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.85 |
| Mean single sequence MFE | -44.48 |
| Consensus MFE | -29.88 |
| Energy contribution | -32.96 |
| Covariance contribution | 3.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7852595 117 - 20766785 UUGCCGUGACCAGGAUUCGAACCUGGGUGUGGUAACCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACAUUGGAUUGC---GCCCAGCAAUUCCAAGAUCCCACCACGUCAC .((.((((.(((((.......))))).(((((....)))))..((((((((........(((.((....)).)))..(((((((((---.....)))).))))))))))))))))).)). ( -42.70) >DroSec_CAF1 10196 119 - 1 UUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUGCAUUGGGUUGCUGGGCGCGGC-UUUCCAAGAUCCCACCACGUCAC ..((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))....(((((..((((....))))-..)))))(((........))).. ( -42.90) >DroSim_CAF1 10646 119 - 1 UUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUGCAUUGGGUUGCUGGGCGCGGC-UUUCCAAGAUCCCACCAGGUCAC ..((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))....(((((..((((....))))-..)))))((((......)))).. ( -45.50) >DroEre_CAF1 10030 119 - 1 UUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACGUCGGGUCGUCGGACGCGCU-CUUCCAAGAUCCCACCAGGUCAC ..((((((.(((((.......))))).....)))))).((...((((((((..........(((((((.((((...)))))))))))(((......-..)))..))))))))...))... ( -47.80) >DroAna_CAF1 10450 119 - 1 UUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACGUCCGACUGUUGGACGAGGC-CGCCUCCGAACCCAGCUGCUCAC .....((((.(((.........(((((((.(((((.......)))))(((....)))......((....(((((.(((((((...))))))).)))-))....)))))))))))).)))) ( -43.50) >consensus UUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACAUUGGGUUGCUGGGCGCGGC_CUUCCAAGAUCCCACCAGGUCAC ..((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))....(((((..((((....))))...)))))((((......)))).. (-29.88 = -32.96 + 3.08)

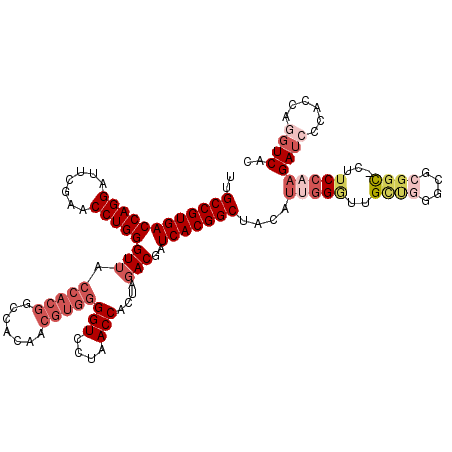
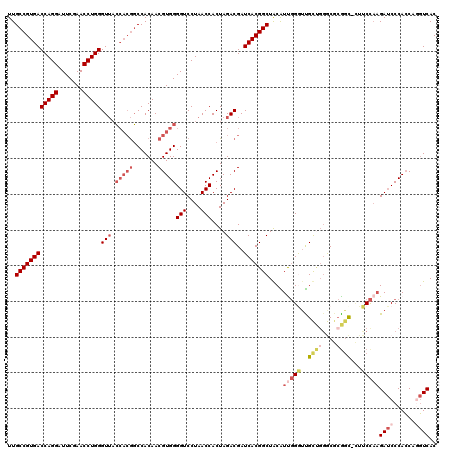
| Location | 7,852,632 – 7,852,751 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.69 |
| Mean single sequence MFE | -40.76 |
| Consensus MFE | -37.14 |
| Energy contribution | -36.74 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7852632 119 + 20766785 CAAUGUAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGGUUACCACACCCAGGUUCGAAUCCUGGUCACGGCAAUGUUCAAAUAAACAUUGUCACGGAGUUGGGUU-AUUUUUU ....((((((((((.......(((((...((((((.....).))))))))))..(((((.......)))))))))(((((((((......))))))))).........))))-))..... ( -41.10) >DroSec_CAF1 10235 119 + 1 CAAUGCAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUUGAAUUAAACAUUGUUACGGAGUUGGGUU-AUAUUUU ....((.((((..((......((((........))))))..)))).)).(((((((((.((((......(....)(((((((((......))))))))).)))).)))))))-))..... ( -38.20) >DroSim_CAF1 10685 119 + 1 CAAUGCAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUUGAAUUAAACAUUGUCACGGAGUUGGGUU-AUAUUUU ....((.((((..((......((((........))))))..)))).)).(((((((((.((((......(....)(((((((((......))))))))).)))).)))))))-))..... ( -40.70) >DroEre_CAF1 10069 116 + 1 CGACGUAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGCUGCGAUAUGCAUUGUCACGGAGUUGGGUU-AA---UU ((((....(((((((......((((........))))......(((((((....(((((.......))))))))))))(((((........)))))))))))).))))....-..---.. ( -41.90) >DroAna_CAF1 10489 120 + 1 GGACGUAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUUGAAACAAACAUUGUCACGGAGUCUGGCUUUCUUUUU ((((....(((((((......((((........))))......(((((((....(((((.......))))))))))))((((((......))))))))))))).))))............ ( -41.90) >consensus CAAUGUAGCCGUGAUCGUCUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGUUGAAAUAAACAUUGUCACGGAGUUGGGUU_AUAUUUU ((((....(((((((......((((........))))......(((((((....(((((.......))))))))))))((((((......))))))))))))).))))............ (-37.14 = -36.74 + -0.40)

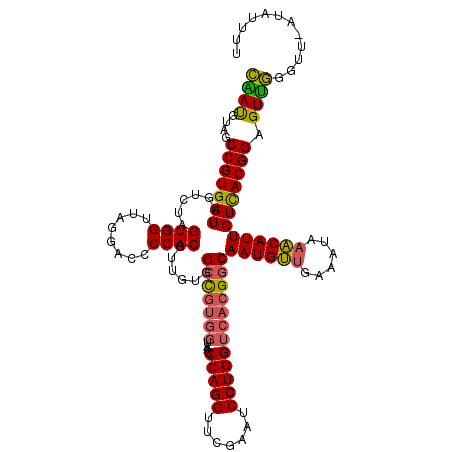
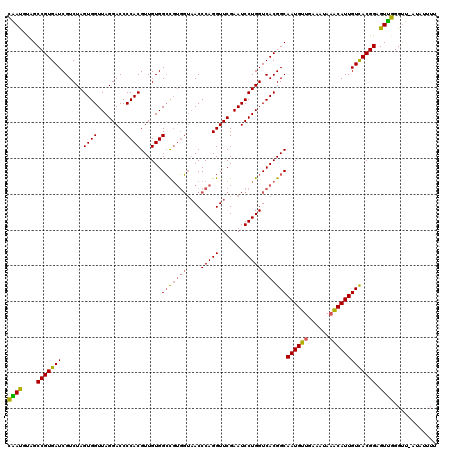
| Location | 7,852,632 – 7,852,751 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.69 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.68 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
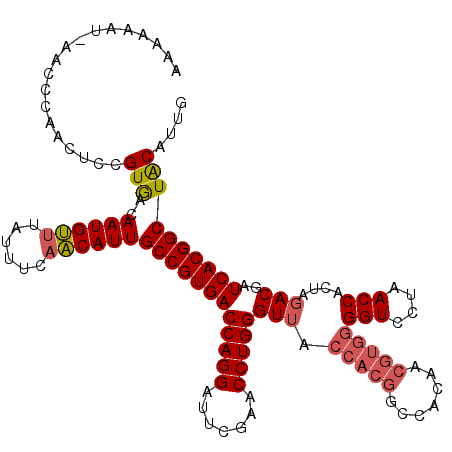
>2R_DroMel_CAF1 7852632 119 - 20766785 AAAAAAU-AACCCAACUCCGUGACAAUGUUUAUUUGAACAUUGCCGUGACCAGGAUUCGAACCUGGGUGUGGUAACCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACAUUG .......-...........(((..(((((((....)))))))((((((((((((.......)))))..(((((..(((((...)))))......))))).......)))))))))).... ( -37.70) >DroSec_CAF1 10235 119 - 1 AAAAUAU-AACCCAACUCCGUAACAAUGUUUAAUUCAACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUGCAUUG .......-...........(((..((((((......))))))((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))))).... ( -35.40) >DroSim_CAF1 10685 119 - 1 AAAAUAU-AACCCAACUCCGUGACAAUGUUUAAUUCAACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUGCAUUG .......-.........((((((...((((((..........((((((.(((((.......))))).....))))))......((((........)))))))))).))))))........ ( -34.60) >DroEre_CAF1 10069 116 - 1 AA---UU-AACCCAACUCCGUGACAAUGCAUAUCGCAGCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACGUCG ..---..-.............((((((((........)))))((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))...))). ( -39.30) >DroAna_CAF1 10489 120 - 1 AAAAAGAAAGCCAGACUCCGUGACAAUGUUUGUUUCAACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACGUCC ........((((.(((((((((..((((((......))))))((((((.(((((.......))))).....)))))).....)))))))))(((.....)))........))))...... ( -37.10) >consensus AAAAAAU_AACCCAACUCCGUGACAAUGUUUAUUUCAACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAGACGAUCACGGCUACAUUG ...................(((..((((((......))))))((((((((((((.......)))))(((.(((((.......)))))(((....)))....)))..)))))))))).... (-31.00 = -31.68 + 0.68)

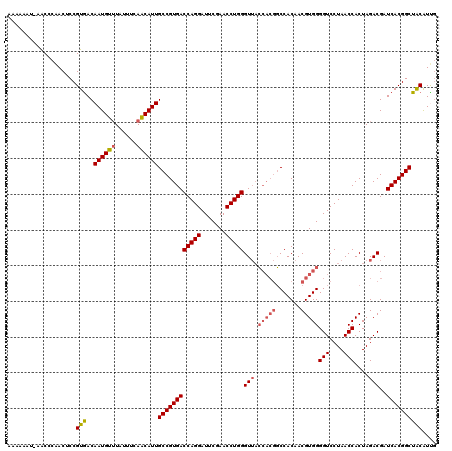
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:12 2006