
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,815,012 – 7,815,124 |
| Length | 112 |
| Max. P | 0.500000 |

| Location | 7,815,012 – 7,815,124 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.82 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -17.57 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7815012 112 + 20766785 UA-UUUUCAAAAAUAUAUAA--CCCAUUUACUCACACUGCCAAAAGUCUUUCCUGGCUGCUGGUAAAUCGUCACCAAAGCCCAGAAGGGCAACUGCAGCACACCCAGUCCCCAGA ..-.................--..............((((.....(((((((..((((..((((........)))).))))..)))))))....))))................. ( -23.40) >DroPse_CAF1 68981 115 + 1 AAGUCUCUUAGGAUGUAUCCCUCCUCCCUACUUACGCUGCUGAAAGUCUUUCCCGGCUGCUGAUAGAUCGUAUAGAUGGCCCAAAUGGGCAGCUGUAGCACUCCCAGGCCCCAGA ..((((....((((((...........(((.(((.((.((((...........)))).))))))))....(((((...((((....))))..)))))))).))).))))...... ( -26.50) >DroSec_CAF1 66309 112 + 1 UA-UUUUCAAAGAUAUAUAA--CCCAUUUACUCACACUGCCAAAAGUCUUUCCUGGCUGCUGGUAAAUCGUGACCAAAGCCCAGAAGGGCAGCUGCAGCACACCCAGUCCCCAGA ..-.....((((((......--.......................)))))).((((..(((((......(((......((((....)))).((....)).))))))))..)))). ( -23.85) >DroSim_CAF1 69039 111 + 1 UA-UUUUCAAAGAUAUAUAA--CCCAGUUACUCACACUGCCAAAAGUCUUUCCUGGCUGCUGGUAAAUCGUGAA-AAAGCCCAGAAGGGCAGCUGCAGCACACCCAGUCCCCAGA ..-.....((((((......--..((((.......))))......)))))).((((..(((((......(((..-...((((....)))).((....)).))))))))..)))). ( -27.04) >DroEre_CAF1 70770 112 + 1 UA-UUUCCAAAGAUAUCUAU--UCUGUUUACUCACGCUGCCAAAAGUCUUGCCUGGCUGCUGGUAAAUCGUGACCAAGGCCCAGAAGGGCAGCUGCAGCACACCCAGUCCCCAGA ..-.................--((((...(((((((.(((((..((((......))))..)))))...))))).....((((....)))).((....)).......))...)))) ( -27.00) >DroAna_CAF1 66279 103 + 1 AA-UUGGAGAGAAAA-----------UAUACUUACACUGGCAAAAGUCUUGCCAGGCUGCUGGUAGAUGGUAUAAACAGCCCAGAAGGGCAGUUGCAACACCCCCAGAGCCCAGA ..-((((........-----------..........(((((((.....)))))))(((.((((..(.((.....(((.((((....)))).)))....)))..))))))))))). ( -33.40) >consensus UA_UUUUCAAAGAUAUAUAA__CCCAUUUACUCACACUGCCAAAAGUCUUUCCUGGCUGCUGGUAAAUCGUGAACAAAGCCCAGAAGGGCAGCUGCAGCACACCCAGUCCCCAGA ...................................(((......))).....((((..(((((...............((((....)))).((....))....)))))..)))). (-17.57 = -18.07 + 0.50)
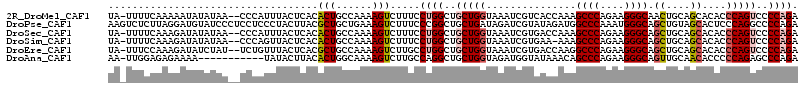
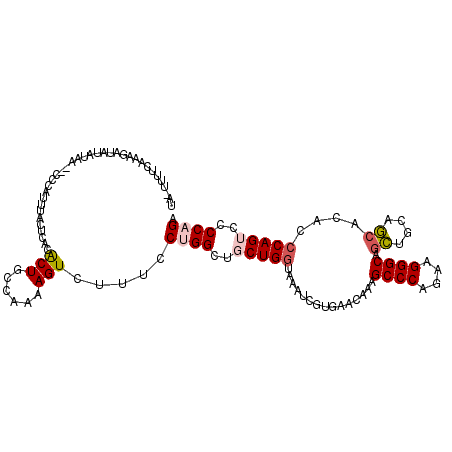
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:47 2006