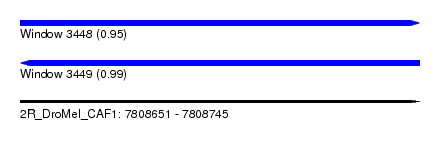
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,808,651 – 7,808,745 |
| Length | 94 |
| Max. P | 0.994398 |

| Location | 7,808,651 – 7,808,745 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -23.52 |
| Energy contribution | -23.40 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
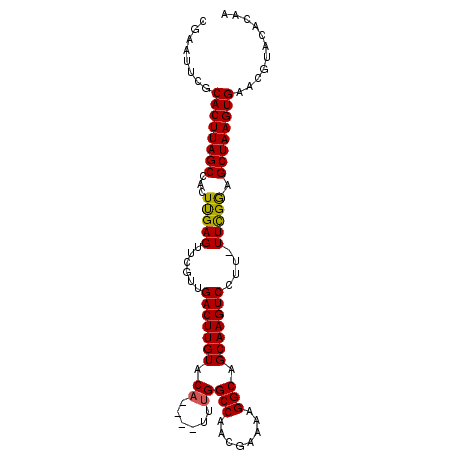
>2R_DroMel_CAF1 7808651 94 + 20766785 UUGUGUACGUUCACUUAGCUCCGAA-AAGGACUUGCUGCCUUUUUGUUGGCCAAA----UGUACAAGUCAACGAACUCAAGAGGCUAAGUGCGAAUUCG .......((..((((((((..((((-((((.(.....))))))))((((((....----.......))))))........)..))))))))))...... ( -25.40) >DroSec_CAF1 59677 94 + 1 UUGUGUACGUUCACUUAGCUCCGAA-AAGGACUUGCUGCCUUUUCGUUGGCCAAA----UGUACAAGUCAACGAACUCAAGUGGCUAAGUGCGAAUUCG .......((..((((((((..((((-((((.(.....))))))))((((((....----.......))))))........)..))))))))))...... ( -28.30) >DroSim_CAF1 62272 95 + 1 UUGUGUACGUUCACUUAGCUUCGAAAAAGGACUUGCUGCCUUUUCGUUGGCCAAA----UGUACAAGUCAACGAACUCAAGUGGCUAAGUGCGAAUUCG .......((..(((((((((.(...(((((.(.....))))))((((((((....----.......))))))))......).)))))))))))...... ( -25.60) >DroEre_CAF1 64120 94 + 1 UUGUGUAUGCUCACUUAGCUCCAAA-AAGGACUUGCUGCCUUUUCGUUGGCCGAA----UGUACAAGUCAACGAACUCAAGGGGCUAAGUGCGAAUUCG .........(.(((((((((((..(-((((.(.....))))))((((((((....----.......))))))))......))))))))))).)...... ( -31.60) >DroYak_CAF1 63578 98 + 1 UUGUGUGUGCUCACUUAGCCCCAAA-AAGGACUUGCUGCCUUUUCGUUGGCCAAAUAUAUGUACAAGUCAACGAACUCGACGGGCUAAGUGUGAAUUCG .........(.((((((((((...(-((((.(.....))))))((((((((...............)))))))).......)))))))))).)...... ( -30.46) >consensus UUGUGUACGUUCACUUAGCUCCGAA_AAGGACUUGCUGCCUUUUCGUUGGCCAAA____UGUACAAGUCAACGAACUCAAGGGGCUAAGUGCGAAUUCG ...........(((((((((.(....((((.(.....)))))(((((((((...............))))))))).....).)))))))))........ (-23.52 = -23.40 + -0.12)

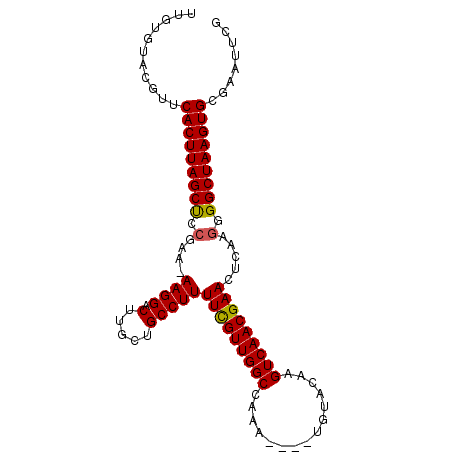
| Location | 7,808,651 – 7,808,745 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -24.10 |
| Energy contribution | -23.74 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7808651 94 - 20766785 CGAAUUCGCACUUAGCCUCUUGAGUUCGUUGACUUGUACA----UUUGGCCAACAAAAAGGCAGCAAGUCCUU-UUCGGAGCUAAGUGAACGUACACAA ........((((((((.(((.(((......(((((((.(.----...)(((........))).)))))))...-))))))))))))))........... ( -25.30) >DroSec_CAF1 59677 94 - 1 CGAAUUCGCACUUAGCCACUUGAGUUCGUUGACUUGUACA----UUUGGCCAACGAAAAGGCAGCAAGUCCUU-UUCGGAGCUAAGUGAACGUACACAA (((((((((.....)).....))))))).......((.((----((((((...(((((((((.....).))))-))))..)))))))).))........ ( -27.50) >DroSim_CAF1 62272 95 - 1 CGAAUUCGCACUUAGCCACUUGAGUUCGUUGACUUGUACA----UUUGGCCAACGAAAAGGCAGCAAGUCCUUUUUCGAAGCUAAGUGAACGUACACAA ........((((((((...(((((......(((((((.(.----...)(((........))).)))))))....))))).))))))))........... ( -24.60) >DroEre_CAF1 64120 94 - 1 CGAAUUCGCACUUAGCCCCUUGAGUUCGUUGACUUGUACA----UUCGGCCAACGAAAAGGCAGCAAGUCCUU-UUUGGAGCUAAGUGAGCAUACACAA ........((((((((.((..(((......(((((((.(.----...)(((........))).))))))))))-...)).))))))))........... ( -25.90) >DroYak_CAF1 63578 98 - 1 CGAAUUCACACUUAGCCCGUCGAGUUCGUUGACUUGUACAUAUAUUUGGCCAACGAAAAGGCAGCAAGUCCUU-UUUGGGGCUAAGUGAGCACACACAA ........((((((((((((((((((....)))))).))..............(((((((((.....).))))-))))))))))))))........... ( -33.00) >consensus CGAAUUCGCACUUAGCCACUUGAGUUCGUUGACUUGUACA____UUUGGCCAACGAAAAGGCAGCAAGUCCUU_UUCGGAGCUAAGUGAACGUACACAA ........((((((((...(((((......(((((((.((......))(((........))).)))))))....))))).))))))))........... (-24.10 = -23.74 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:46 2006