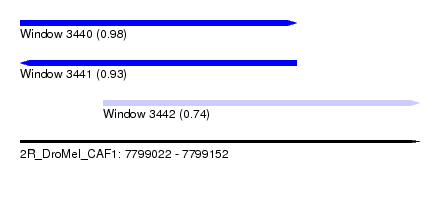
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,799,022 – 7,799,152 |
| Length | 130 |
| Max. P | 0.982215 |

| Location | 7,799,022 – 7,799,112 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 73.28 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.33 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7799022 90 + 20766785 CUCCGGAGAAAAGGAUAUAUGUAUAUAUAUGUGUCCUAG-----------------AUGUUUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUG ...((((....(((((((((((....))))))))))).(-----------------((....(((((.........)))))((((((...)))))).))).)))).. ( -28.10) >DroSec_CAF1 50129 86 + 1 UUCCGGAGAAAAGGAUAUAUGUG----UAUGUGUCCUAG-----------------AUGUUUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUA ...((((....(((((((((...----.))))))))).(-----------------((....(((((.........)))))((((((...)))))).))).)))).. ( -25.40) >DroSim_CAF1 52206 86 + 1 UUCCGGAGAAAAGGAUAUAUGUG----UAUGUGUCCUAG-----------------AUGUUUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUA ...((((....(((((((((...----.))))))))).(-----------------((....(((((.........)))))((((((...)))))).))).)))).. ( -25.40) >DroEre_CAF1 54513 80 + 1 CUCCAGAGAAAUGGACAUAG----------AUGUCCUAG-----------------AUGCAUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUA ..((((......(((((...----------.)))))...-----------------((((.....))))........))))((((((...))))))........... ( -22.40) >DroYak_CAF1 54061 80 + 1 CUCCAGAGGAAAGGACACGG----------AUGUCCUAG-----------------AUGUAUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCACUCGUA .....(((...((((((...----------.)))))).(-----------------((....(((((.........)))))((((((...)))))).))).)))... ( -25.20) >DroAna_CAF1 46831 99 + 1 UUCCUUAAGAAAGGAGGAAUAU--------CUUUUUUGGAUUUUUUUUUUAUGGCCAUGCAUCGAGUAUAGUUACAGCCAUUUGGCCAAAGGUCGCUGUCAGUCGAU .(((...((((((((......)--------))))))))))....................(((((.((((((.((.(((....))).....)).))))).).))))) ( -20.30) >consensus CUCCGGAGAAAAGGAUAUAUGU________GUGUCCUAG_________________AUGUAUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUA ..((((.....(((((((............)))))))............................(((....)))..))))((((((...))))))........... (-16.66 = -16.33 + -0.33)

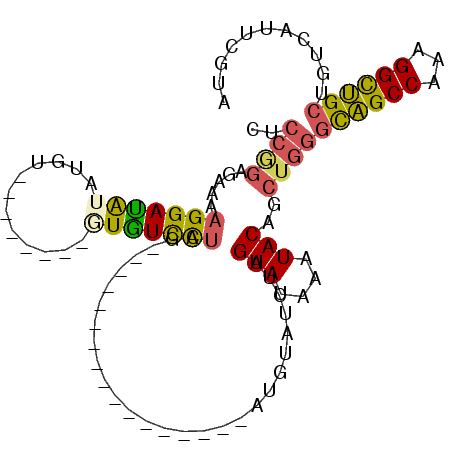
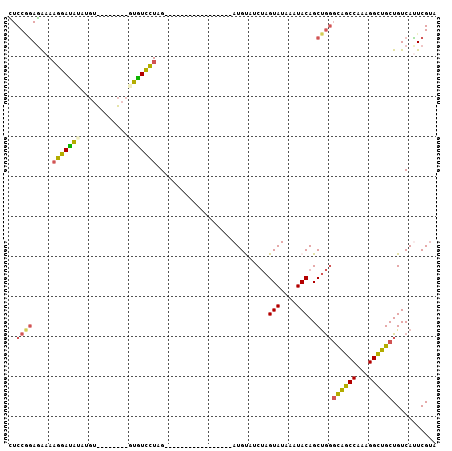
| Location | 7,799,022 – 7,799,112 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 73.28 |
| Mean single sequence MFE | -20.63 |
| Consensus MFE | -10.53 |
| Energy contribution | -10.42 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7799022 90 - 20766785 CACGAAUGACAGCAGCCUUUGGCUGCCCAGCUGUAUUUAUACUAGAAACAU-----------------CUAGGACACAUAUAUAUACAUAUAUCCUUUUCUCCGGAG ...........((((((...)))))).....(((((.(((((((((....)-----------------))))......)))).)))))....(((........))). ( -18.50) >DroSec_CAF1 50129 86 - 1 UACGAAUGACAGCAGCCUUUGGCUGCCCAGCUGUAUUUAUACUAGAAACAU-----------------CUAGGACACAUA----CACAUAUAUCCUUUUCUCCGGAA .....(((...((((((...)))))).....(((((.....(((((....)-----------------)))).....)))----)))))...(((........))). ( -19.70) >DroSim_CAF1 52206 86 - 1 UACGAAUGACAGCAGCCUUUGGCUGCCCAGCUGUAUUUAUACUAGAAACAU-----------------CUAGGACACAUA----CACAUAUAUCCUUUUCUCCGGAA .....(((...((((((...)))))).....(((((.....(((((....)-----------------)))).....)))----)))))...(((........))). ( -19.70) >DroEre_CAF1 54513 80 - 1 UACGAAUGACAGCAGCCUUUGGCUGCCCAGCUGUAUUUAUACUAGAUGCAU-----------------CUAGGACAU----------CUAUGUCCAUUUCUCUGGAG ...........((((((...))))))((((.((((((((...)))))))).-----------------...(((((.----------...)))))......)))).. ( -24.90) >DroYak_CAF1 54061 80 - 1 UACGAGUGACAGCAGCCUUUGGCUGCCCAGCUGUAUUUAUACUAGAUACAU-----------------CUAGGACAU----------CCGUGUCCUUUCCUCUGGAG ((((..((...((((((...))))))((((.((((((((...)))))))).-----------------)).)).)).----------.))))(((........))). ( -25.30) >DroAna_CAF1 46831 99 - 1 AUCGACUGACAGCGACCUUUGGCCAAAUGGCUGUAACUAUACUCGAUGCAUGGCCAUAAAAAAAAAAUCCAAAAAAG--------AUAUUCCUCCUUUCUUAAGGAA .(((........)))....((((((....((.(((....))).....)).)))))).....................--------.......(((((....))))). ( -15.70) >consensus UACGAAUGACAGCAGCCUUUGGCUGCCCAGCUGUAUUUAUACUAGAAACAU_________________CUAGGACAC________ACAUAUAUCCUUUUCUCCGGAA ........(((((((((...)))).....)))))..........................................................(((........))). (-10.53 = -10.42 + -0.11)

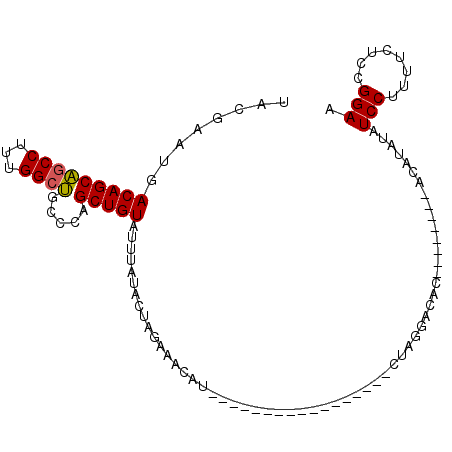
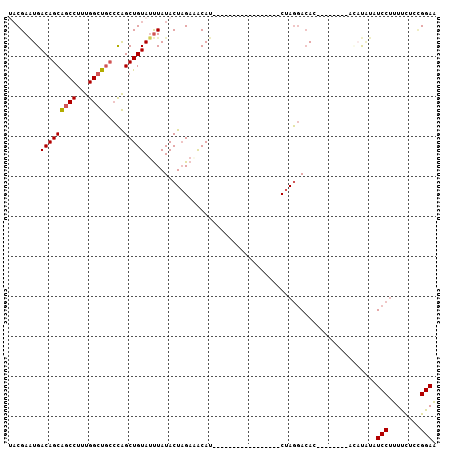
| Location | 7,799,049 – 7,799,152 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -18.65 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
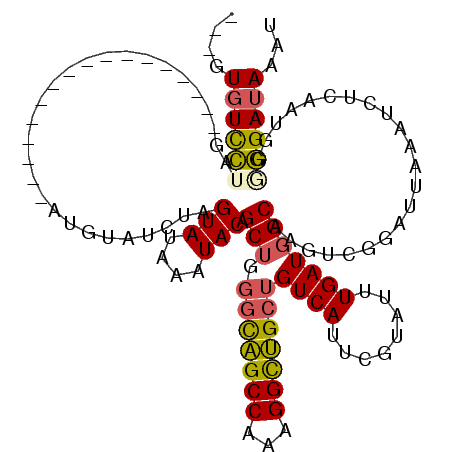
>2R_DroMel_CAF1 7799049 103 + 20766785 UAUGUGUCCUAG-----------------AUGUUUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUGUUUGAUGGCAAGUUGGAUUAAAUCUCGAUGGGGGAUAAAU (((.(((.((((-----------------(....))))).))).)))(((((.(((((((...)))))))(((((.((....))))))).)))))......(((((.....))))).... ( -29.70) >DroSec_CAF1 50152 103 + 1 UAUGUGUCCUAG-----------------AUGUUUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUAUUUGAUGGCAAGCUGGAUUAAAUCUCAAUGGGGGAUAAAU (((.(((.((((-----------------(....))))).))).)))(((((.(((((((...)))))))(((((.((....))))))).)))))......(((((.....))))).... ( -31.90) >DroSim_CAF1 52229 103 + 1 UAUGUGUCCUAG-----------------AUGUUUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUAUUUGAUGGCAAGUUGGAUUAAAUCUCAAUGGGGGAUAAAU (((.(((.((((-----------------(....))))).))).)))(((((.(((((((...)))))))(((((.((....))))))).)))))......(((((.....))))).... ( -29.50) >DroEre_CAF1 54533 99 + 1 ---AUGUCCUAG-----------------AUGCAUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUAUUUGAUGGCAAGUCGGAUUAAAUCUCAAUGG-GGAUAAAU ---.((((((.(-----------------((....(((((.........)))))((((((...)))))).)))(((((.(((((....))))))))))............)-)))))... ( -26.80) >DroYak_CAF1 54081 99 + 1 ---AUGUCCUAG-----------------AUGUAUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCACUCGUAUUUGAUGGCAAGUCGGAUUAAAUCUCAAUGG-GGAUAAAU ---.((((((((-----------------((..((((.(((....))).(((.(((((((...)))))))((((.(((....))))))).))).))))...)))).....)-)))))... ( -27.20) >DroAna_CAF1 46853 117 + 1 ---CUUUUUUGGAUUUUUUUUUUAUGGCCAUGCAUCGAGUAUAGUUACAGCCAUUUGGCCAAAGGUCGCUGUCAGUCGAUGCUGAUUGCCAGUCGGAUUAAAUCUCAGGGGAAGAUAAAU ---.((((((((((((..(((...((((...(((((((.((((((.((.(((....))).....)).))))).).))))))).....))))...)))..))))).)))))))........ ( -34.80) >consensus ___GUGUCCUAG_________________AUGUAUCUAGUAUAAAUACAGCUGGGCAGCCAAAGGCUGCUGUCAUUCGUAUUUGAUGGCAAGUCGGAUUAAAUCUCAAUGGGGGAUAAAU ....((((((............................(((....))).(((.(((((((...)))))))((((........)))))))......................))))))... (-18.65 = -18.60 + -0.05)

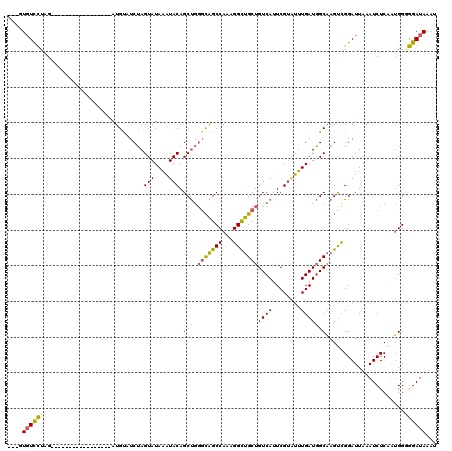
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:39 2006