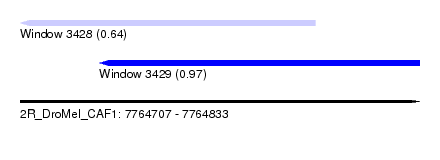
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,764,707 – 7,764,833 |
| Length | 126 |
| Max. P | 0.974715 |

| Location | 7,764,707 – 7,764,800 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -16.44 |
| Energy contribution | -17.42 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
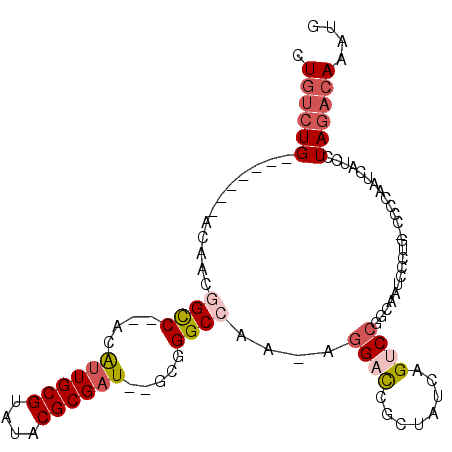
>2R_DroMel_CAF1 7764707 93 - 20766785 CUGUCUG-------GUCAGGGCC--ACAUUGCGUAUACGCGAU--GCGGGCCAA-AGGACCGCUAUCAGUCCGGCAAUUCCUG-UCCCAAUCAUCCUAGACAAAUG .((((((-------(...((.((--.(((((((....))))))--).)).))..-.((((........))))((((.....))-)).........))))))).... ( -34.50) >DroPse_CAF1 20916 85 - 1 CUGUCUG-------ACAGCAGCC--ACAUUGCGUAUACGCGAU--GCGGGCGA---GG-UCG-----UGUCCCGCCAUCUAUU-CCCCAAUCAUCCUAGACAAAUG .((((((-------........(--.(((((((....))))))--).)((((.---((-...-----...)))))).......-............)))))).... ( -25.70) >DroEre_CAF1 19861 94 - 1 CUGUCUG-------GCCAAGGCC--ACAUUGCGUAUACGCGAU--GCGGGCCAAAAGGACCGCUAUCAGUCCGGCCAUUCCUG-UCCCAAUCAUCCUAGACAAAUG .((((((-------(((..((((--.(((((((....))))))--)..))))....((((........)))))))).....((-.......))....))))).... ( -35.50) >DroYak_CAF1 18332 94 - 1 CUGUGUG-------GCCAAGGCC--ACGUUGCGUAUACGCGAU--GCGGGCCAAAAGGACCGGUAUCAGUCCGGCAAUCCCUG-UCCCAAUCAUCCUAGACAAAUG ....(((-------((....)))--))((((((....)))(((--..(((.((...(((((((.......))))...))).))-.))).)))......)))..... ( -30.80) >DroAna_CAF1 15827 106 - 1 CUCCCUGCCGUCCGACAGCGGUCAUACAGUGCGUAUACGCGAUGUGCGGGCCAAAAGGAUCGCCAUCAGUCGGCCAAUCUCUGGCCCCAAUCAUCCUAGACAAAUG .........(((.(((.(((...((((.....)))).)))((((.(((..((....))..))))))).)))(((((.....)))))............)))..... ( -33.90) >DroPer_CAF1 20302 85 - 1 CUGUCUG-------ACAGCAGCC--ACAUUGCGUAUACGCGAU--GCGGGCGA---GG-UCG-----UGUCCCGCCAUCUAUU-CCCCAAUCAUCCUAGACAAAUG .((((((-------........(--.(((((((....))))))--).)((((.---((-...-----...)))))).......-............)))))).... ( -25.70) >consensus CUGUCUG_______ACAACGGCC__ACAUUGCGUAUACGCGAU__GCGGGCCAA_AGGACCGCUAUCAGUCCGGCAAUCCCUG_CCCCAAUCAUCCUAGACAAAUG .((((((............((((....((((((....)))))).....))))....((((........))))........................)))))).... (-16.44 = -17.42 + 0.97)

| Location | 7,764,732 – 7,764,833 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -13.81 |
| Energy contribution | -13.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7764732 101 - 20766785 UAAUCAUCCUUGGCCACAAACUUUUCCACUUAACUGUCUG-------GUCAGGGCC--ACAUUGCGUAUACGCGAU--GCGGGCCAA-AGGACCGCUAUCAGUCCGGCAAUUC .......(((((((((...((..............)).))-------)))))))((--.(((((((....))))))--).))(((..-.((((........)))))))..... ( -32.94) >DroPse_CAF1 20941 90 - 1 CAAUCU---ACGACCACAAACUUUUCCACUCAACUGUCUG-------ACAGCAGCC--ACAUUGCGUAUACGCGAU--GCGGGCGA---GG-UCG-----UGUCCCGCCAUCU .....(---((((((..................(((....-------.)))..(((--.(((((((....))))))--)..)))..---))-)))-----))........... ( -27.40) >DroEre_CAF1 19886 102 - 1 UAAUCAUCCUUGGCCACAAACUUUUCCACUUAACUGUCUG-------GCCAAGGCC--ACAUUGCGUAUACGCGAU--GCGGGCCAAAAGGACCGCUAUCAGUCCGGCCAUUC .......(((((((((...((..............)).))-------))))))).(--.(((((((....))))))--).)((((....((((........)))))))).... ( -35.64) >DroYak_CAF1 18357 102 - 1 UAAUCAUCCUUGGCCACAAACUUUUCCACUUAACUGUGUG-------GCCAAGGCC--ACGUUGCGUAUACGCGAU--GCGGGCCAAAAGGACCGGUAUCAGUCCGGCAAUCC ......((((((((((((...((........))...))))-------)))..((((--.(((((((....))))))--)..))))..)))))((((.......))))...... ( -40.20) >DroAna_CAF1 15853 111 - 1 UAAUCAA--AUGACAAGAAACUCUUCCACUCAACUCCCUGCCGUCCGACAGCGGUCAUACAGUGCGUAUACGCGAUGUGCGGGCCAAAAGGAUCGCCAUCAGUCGGCCAAUCU .......--.(((...(((....)))...)))..........(.(((((.(((...((((.....)))).)))((((.(((..((....))..))))))).))))).)..... ( -28.70) >DroPer_CAF1 20327 90 - 1 CAAUCU---ACGACCACAAACUUUUCCACUCAACUGUCUG-------ACAGCAGCC--ACAUUGCGUAUACGCGAU--GCGGGCGA---GG-UCG-----UGUCCCGCCAUCU .....(---((((((..................(((....-------.)))..(((--.(((((((....))))))--)..)))..---))-)))-----))........... ( -27.40) >consensus UAAUCAU__AUGACCACAAACUUUUCCACUCAACUGUCUG_______ACAACGGCC__ACAUUGCGUAUACGCGAU__GCGGGCCAA_AGGACCGCUAUCAGUCCGGCAAUCC ..........(((((............((......))...............((((....((((((....)))))).....))))....)).))).................. (-13.81 = -13.82 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:27 2006