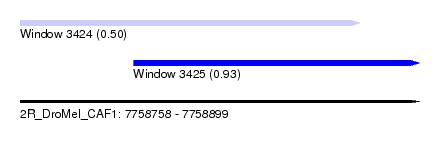
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,758,758 – 7,758,899 |
| Length | 141 |
| Max. P | 0.927972 |

| Location | 7,758,758 – 7,758,878 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.77 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -22.72 |
| Energy contribution | -23.75 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
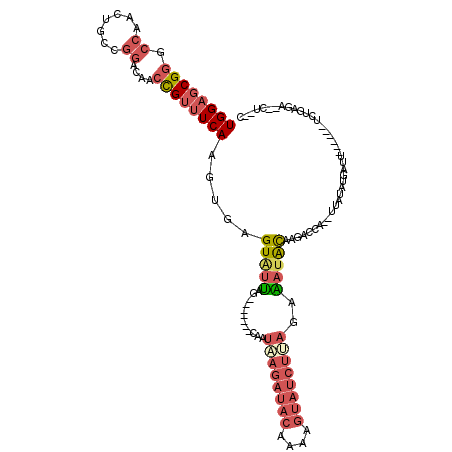
>2R_DroMel_CAF1 7758758 120 + 20766785 UCCACACGCAGGUGUCGUCGGUUCAGGAACAGAUGCCUCAUGGAGCGGGCCAACUGCCGGACAACCGUUUCAAGUGAGUAUUAGCUGAAUGCAAAAAGAUACAAAAGUAUCUUAGAAAUA ....((((.(((((((....(((....))).)))))))).((((((((.((.......))....)))))))).))).(((((.....)))))...(((((((....)))))))....... ( -33.70) >DroPse_CAF1 15181 90 + 1 AGGACACGCCGGUGCCACCGGUUCAAGAACCGCUGCCUAAUGGAGCGAGCCAAUUGCCGGACAAGUGUCUCGAGUGAGUCUCAA-G-----GAAUA------------------------ .((((((.((((((((...((((....))))(((.((....)))))).)).....)))))....)))))).(((.....)))..-.-----.....------------------------ ( -29.30) >DroSec_CAF1 10738 113 + 1 UCCACACGCAGGUGUCGUCGGUUUAGGAACAGGUGCCUCUUGGAGCGGGCCAACUGCCGGACGACCGUUUCAAGUGAGUGUUAG-------CAAUAAGAUACAAAAGUAUCUUAGAAAUA ...((((.((((..((.((.......))...))..)).((((((((((.((.......))....)))))))))))).))))...-------...((((((((....))))))))...... ( -34.10) >DroSim_CAF1 11526 113 + 1 UCCACACGCAGGUGUCGUCGGUUUAGGAACAGGUGCCUCUUGGAGCGGGCCAACUGCCGGACGACCGUUUCAAGUGAGUGUUAG-------CAAUAAGAUACAAAAGUAUCUUAGAAAUA ...((((.((((..((.((.......))...))..)).((((((((((.((.......))....)))))))))))).))))...-------...((((((((....))))))))...... ( -34.10) >DroEre_CAF1 14239 120 + 1 UCCACUCGCAGGUGUCAACGGUUCAAGAACAGGUGCCUCAUGGAGCGGGCCAACUGCCGGACAACCGUUUCAAGUGAGUAUAGGCUGAAUCCAAUGAGAUACAAAAGUAUCUCAGAUAUG ...(((((((((..((....(((....))).))..)))..((((((((.((.......))....)))))))).))))))...((......))..((((((((....))))))))...... ( -37.70) >DroYak_CAF1 12719 120 + 1 UCCAGUCGCAGGUGUCAUCGGUUCAAGAACAGGUGCCUCAUGGAGCGGGCCAACUGCCGGACAACCGUUUCAAGUGAGUAUUGGCUGAAUCCCAUUAGAUACAAAAGUAUCUUAGAAAUG ..((((((((..(((..((.......)))))..)))((((((((((((.((.......))....))))))))..))))....))))).........((((((....))))))........ ( -31.10) >consensus UCCACACGCAGGUGUCAUCGGUUCAAGAACAGGUGCCUCAUGGAGCGGGCCAACUGCCGGACAACCGUUUCAAGUGAGUAUUAG_U_____CAAUAAGAUACAAAAGUAUCUUAGAAAUA ...((.(((.((..((....(((....))).))..))...((((((((.((.......))....)))))))).))).))...............((((((((....))))))))...... (-22.72 = -23.75 + 1.03)



| Location | 7,758,798 – 7,758,899 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.26 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -13.32 |
| Energy contribution | -14.66 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7758798 101 + 20766785 UGGAGCGGGCCAACUGCCGGACAACCGUUUCAAGUGAGUAUUAGCUGAAUGCAAAAAGAUACAAAAGUAUCUUAGAAAUACAAGACCA-UUUAUAUGAUU------UC------------ ((((((((.((.......))....)))))))).....(((((.((.....))...(((((((....)))))))...))))).......-...........------..------------ ( -23.20) >DroSec_CAF1 10778 101 + 1 UGGAGCGGGCCAACUGCCGGACGACCGUUUCAAGUGAGUGUUAG-------CAAUAAGAUACAAAAGUAUCUUAGAAAUACAAGAC------AUAUGCUG------UUUGACUUUUUUAC ((((((((.((.......))....)))))))).(((((((((..-------...((((((((....))))))))..))))).((((------(.....))------))).......)))) ( -24.40) >DroSim_CAF1 11566 104 + 1 UGGAGCGGGCCAACUGCCGGACGACCGUUUCAAGUGAGUGUUAG-------CAAUAAGAUACAAAAGUAUCUUAGAAAUACAAGACCA-UUUAUAUGAUU------UUUGAGA--CUUAC ((((((((.((.......))....)))))))).((((((.((((-------...((((((((....))))))))............((-(....)))...------.)))).)--))))) ( -27.30) >DroEre_CAF1 14279 102 + 1 UGGAGCGGGCCAACUGCCGGACAACCGUUUCAAGUGAGUAUAGGCUGAAUCCAAUGAGAUACAAAAGUAUCUCAGAUAUGCCAGAA----------------CGUAUCUCAGA--CUAGC ((((((((.((.......))....))))))))........(((.((((......((((((((....))))))))((((((......----------------)))))))))).--))).. ( -32.60) >DroYak_CAF1 12759 117 + 1 UGGAGCGGGCCAACUGCCGGACAACCGUUUCAAGUGAGUAUUGGCUGAAUCCCAUUAGAUACAAAAGUAUCUUAGAAAUGCCAGAAUA-CUUGUCUGAAUUACAUAUCUGAGA--CUAGC ((((((((.((.......))....))))))))...((((((((((....((.....((((((....))))))..))...))))..)))-)))((((.(..........).)))--).... ( -28.50) >DroAna_CAF1 10523 83 + 1 UGGAACGAACCAAUUGCAAUGCAAGUGGAUCAAGUGAGUCUCAA-------AAAUAGUCUAUA----------AGGGUUAUGAGACCAAGUUAUAUAAUA-------------------- ...(((((.(((.((((...)))).))).))...((.((((((.-------...((..((...----------.))..)))))))))).)))........-------------------- ( -17.80) >consensus UGGAGCGGGCCAACUGCCGGACAACCGUUUCAAGUGAGUAUUAG_______CAAUAAGAUACAAAAGUAUCUUAGAAAUACAAGACCA__UUAUAUGAUU______UCUGAGA__CU__C ((((((((.((.......))....)))))))).....(((((............((((((((....))))))))..)))))....................................... (-13.32 = -14.66 + 1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:23 2006