
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,730,727 – 7,730,818 |
| Length | 91 |
| Max. P | 0.808285 |

| Location | 7,730,727 – 7,730,818 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 81.04 |
| Mean single sequence MFE | -16.75 |
| Consensus MFE | -11.48 |
| Energy contribution | -11.20 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
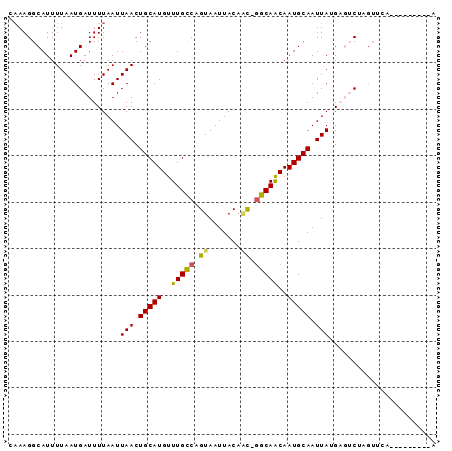
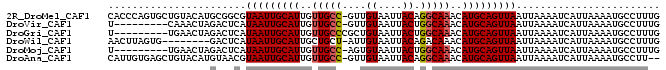
>2R_DroMel_CAF1 7730727 91 + 20766785 CAAAGGCAUUUUAAUGAUUUUAAUUAACUGCAUGUUUGCCUGUAAUUACAAC-GGCAACAAUGCAAUUACGCCGCAUGUACAGCACUGGGUG ....(((...(((((.......))))).(((((..((((((((....)))..-)))))..))))).....)))((.......))........ ( -19.20) >DroVir_CAF1 13227 82 + 1 CAAAGGCAUUUUAAUGAUUUUAAUUAACUGCAUGUUUGCCAGUAAUUACAAC-GGCAACAAUGCAAUUAUGAGUCUAGUUUG---------A ......((..(((..(((((....(((.(((((..(((((.((.......))-)))))..))))).))).))))))))..))---------. ( -15.70) >DroGri_CAF1 15308 83 + 1 CAAAGGCAUUUUAAUGAUUUUAAUUAACUGCAUGUUUGCCAGUAAUUACAGCGGGCAACAAUGCAAUUAUGAGUCUAGUUCA---------A ...((((...(((((.......))))).(((((..(((((.((.......)).)))))..))))).......))))......---------. ( -16.90) >DroWil_CAF1 14399 83 + 1 CAAAGGCAUUUUAAUGAUUUUAAUUAACUGCAUGUUUGUCUGUAAUUACAAU-AGCAGCAAUGCAAUUAUGAGUC--------CACUAAGUU ....((.((((..((((((.((((((...(((....)))...))))))....-.(((....))))))))))))))--------)........ ( -11.90) >DroMoj_CAF1 15008 82 + 1 CAAAGGCAUUUUAAUGAUUUUAAUUAACUGCAUGUUUGCCAGUAAUUACACU-GGCAACAAUGCAAUUAUGAGUCUAGUUCA---------A ...((((...(((((.......))))).(((((..((((((((......)))-)))))..))))).......))))......---------. ( -21.50) >DroAna_CAF1 11345 89 + 1 --AAGGCAUUUUAAUGAUUUUAAUUAACUGCAUGUUUGCCUGUAAUUACAAC-GGCAACAAUGCAAUUACGUUACAUGUACAGCUCACAAUG --..(((.....................(((((..((((((((....)))..-)))))..)))))..(((((...)))))..)))....... ( -15.30) >consensus CAAAGGCAUUUUAAUGAUUUUAAUUAACUGCAUGUUUGCCAGUAAUUACAAC_GGCAACAAUGCAAUUAUGAGUCUAGUUCA_________A ........................(((.(((((..(((((.((.......)).)))))..))))).)))....................... (-11.48 = -11.20 + -0.28)


| Location | 7,730,727 – 7,730,818 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 81.04 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -13.20 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
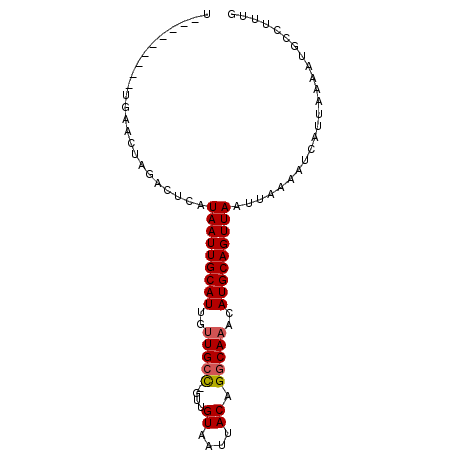
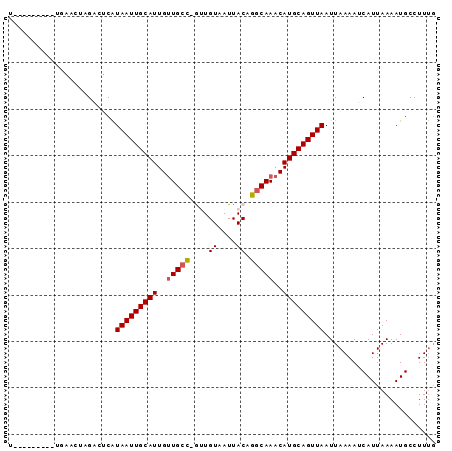
>2R_DroMel_CAF1 7730727 91 - 20766785 CACCCAGUGCUGUACAUGCGGCGUAAUUGCAUUGUUGCC-GUUGUAAUUACAGGCAAACAUGCAGUUAAUUAAAAUCAUUAAAAUGCCUUUG ........(((((....))))).(((((((((..(((((-..(((....))))))))..)))))))))........................ ( -22.50) >DroVir_CAF1 13227 82 - 1 U---------CAAACUAGACUCAUAAUUGCAUUGUUGCC-GUUGUAAUUACUGGCAAACAUGCAGUUAAUUAAAAUCAUUAAAAUGCCUUUG .---------.............(((((((((..(((((-(.((....)).))))))..)))))))))........................ ( -16.90) >DroGri_CAF1 15308 83 - 1 U---------UGAACUAGACUCAUAAUUGCAUUGUUGCCCGCUGUAAUUACUGGCAAACAUGCAGUUAAUUAAAAUCAUUAAAAUGCCUUUG .---------.............(((((((((..(((((.(.((....))).)))))..)))))))))........................ ( -15.20) >DroWil_CAF1 14399 83 - 1 AACUUAGUG--------GACUCAUAAUUGCAUUGCUGCU-AUUGUAAUUACAGACAAACAUGCAGUUAAUUAAAAUCAUUAAAAUGCCUUUG ...((((((--------(.....((((((((((((((.(-(.......))))).))...))))))))).......))))))).......... ( -12.60) >DroMoj_CAF1 15008 82 - 1 U---------UGAACUAGACUCAUAAUUGCAUUGUUGCC-AGUGUAAUUACUGGCAAACAUGCAGUUAAUUAAAAUCAUUAAAAUGCCUUUG .---------.............(((((((((..(((((-((((....)))))))))..)))))))))........................ ( -23.60) >DroAna_CAF1 11345 89 - 1 CAUUGUGAGCUGUACAUGUAACGUAAUUGCAUUGUUGCC-GUUGUAAUUACAGGCAAACAUGCAGUUAAUUAAAAUCAUUAAAAUGCCUU-- ((((((((((.......))....(((((((((..(((((-..(((....))))))))..))))))))).......))))...))))....-- ( -17.80) >consensus U_________UGAACUAGACUCAUAAUUGCAUUGUUGCC_GUUGUAAUUACAGGCAAACAUGCAGUUAAUUAAAAUCAUUAAAAUGCCUUUG .......................(((((((((..(((((....((....)).)))))..)))))))))........................ (-13.20 = -13.40 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:13 2006