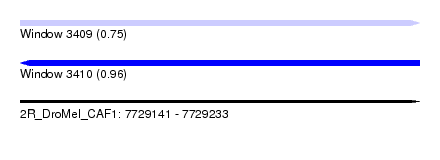
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,729,141 – 7,729,233 |
| Length | 92 |
| Max. P | 0.956807 |

| Location | 7,729,141 – 7,729,233 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
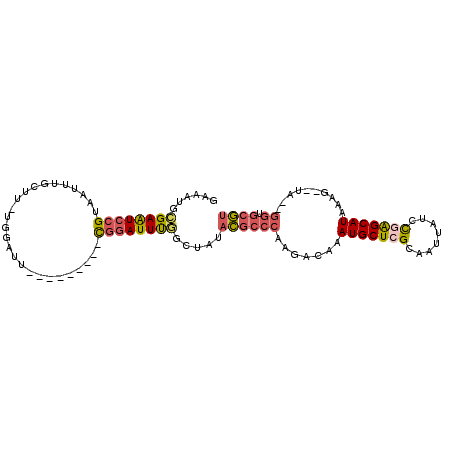
>2R_DroMel_CAF1 7729141 92 + 20766785 GAAAUGAGAAUCCGUAAUUUGCUUUUGGAUA---------CGGAUUUGGCUGUACGCCCAAGACAAAUGCUCGCAAUUAUCCGAGCAUAAAG--UA--GGUGCGU .......(((((((((.(((......)))))---------)))))))......((((((...((..(((((((........)))))))...)--).--)).)))) ( -25.80) >DroSec_CAF1 19111 91 + 1 GAAAUGCGAAUCCGUAAUUUGCUU-UGGAUU---------CGGAUUUGGCUCUACGCCCAAGACAAAUGCUCGCAAUUAUCCGAGCAUAAAG--UA--GGUGCGU .((((.((((((((..........-))))))---------)).))))......((((((...((..(((((((........)))))))...)--).--)).)))) ( -29.00) >DroSim_CAF1 17483 91 + 1 GAAAUGCGAAUCCGUAAUUUGCUU-UGGAUU---------CGGAUUUGGCUCUACGCCCAAGACAAAUGCUCGCAAUUAUCCGAGCAUAAAG--UA--GGUGCGU .((((.((((((((..........-))))))---------)).))))......((((((...((..(((((((........)))))))...)--).--)).)))) ( -29.00) >DroEre_CAF1 12580 91 + 1 GGCAUCCGAAUCCGUAAUUUGGUA-UGGAUU---------UGGAUUUGGCUAUACGCCCAAGACAAAUGCUCGCAAUUAUCCAGGCAUAAAG--UA--GGUGCGU ((((((((((((((((......))-))))))---------)))))...)))..((((((...((..((((..(........)..))))...)--).--)).)))) ( -28.40) >DroYak_CAF1 12977 91 + 1 GAAAUCCGAGUCCGUAAUUUGGCU-UGAAUU---------UGGAUUCGGCUGUACGCCCAAGACAAAUGCUCGCAAUUAUCCGAGCAUAAAG--UG--GGUGCGU .....(((((((((.(((((....-.)))))---------))))))))).....((((((......(((((((........)))))))....--))--))))... ( -32.80) >DroAna_CAF1 9736 104 + 1 GAAAUUGGAAUUGGUAGCUUAGUU-UAUAUUGUAGAUACAUACAUUUUAUUAUCUGGGAAAGACAAAUGCUAGCAAUUAUUUGGGCAUAUAAGCCAGGGGUGGAA ........(((((.((((((.(((-(...((.((((((.(((.....))))))))).)).)))).)).)))).))))).....(((......))).......... ( -19.70) >consensus GAAAUGCGAAUCCGUAAUUUGCUU_UGGAUU_________CGGAUUUGGCUAUACGCCCAAGACAAAUGCUCGCAAUUAUCCGAGCAUAAAG__UA__GGUGCGU ......((((((((..........................)))))))).....((((((.......(((((((........)))))))..........)).)))) (-14.22 = -14.50 + 0.28)

| Location | 7,729,141 – 7,729,233 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -12.27 |
| Energy contribution | -12.33 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
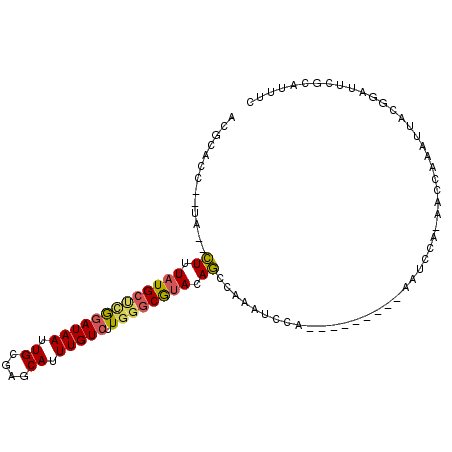
>2R_DroMel_CAF1 7729141 92 - 20766785 ACGCACC--UA--CUUUAUGCUCGGAUAAUUGCGAGCAUUUGUCUUGGGCGUACAGCCAAAUCCG---------UAUCCAAAAGCAAAUUACGGAUUCUCAUUUC ((((.((--.(--(...(((((((.(....).)))))))..))...)))))).......((((((---------((.............))))))))........ ( -26.72) >DroSec_CAF1 19111 91 - 1 ACGCACC--UA--CUUUAUGCUCGGAUAAUUGCGAGCAUUUGUCUUGGGCGUAGAGCCAAAUCCG---------AAUCCA-AAGCAAAUUACGGAUUCGCAUUUC ..((...--..--((((((((((((((((.((....)).))))).)))))))))))........(---------(((((.-...........))))))))..... ( -28.80) >DroSim_CAF1 17483 91 - 1 ACGCACC--UA--CUUUAUGCUCGGAUAAUUGCGAGCAUUUGUCUUGGGCGUAGAGCCAAAUCCG---------AAUCCA-AAGCAAAUUACGGAUUCGCAUUUC ..((...--..--((((((((((((((((.((....)).))))).)))))))))))........(---------(((((.-...........))))))))..... ( -28.80) >DroEre_CAF1 12580 91 - 1 ACGCACC--UA--CUUUAUGCCUGGAUAAUUGCGAGCAUUUGUCUUGGGCGUAUAGCCAAAUCCA---------AAUCCA-UACCAAAUUACGGAUUCGGAUGCC ..((...--..--((.(((((((((((((.((....)).))))).)))))))).))....((((.---------(((((.-((......)).))))).)))))). ( -26.30) >DroYak_CAF1 12977 91 - 1 ACGCACC--CA--CUUUAUGCUCGGAUAAUUGCGAGCAUUUGUCUUGGGCGUACAGCCGAAUCCA---------AAUUCA-AGCCAAAUUACGGACUCGGAUUUC ((((.((--.(--(...(((((((.(....).)))))))..))...))))))....((((.(((.---------......-...........))).))))..... ( -25.97) >DroAna_CAF1 9736 104 - 1 UUCCACCCCUGGCUUAUAUGCCCAAAUAAUUGCUAGCAUUUGUCUUUCCCAGAUAAUAAAAUGUAUGUAUCUACAAUAUA-AACUAAGCUACCAAUUCCAAUUUC ..........(((......))).....(((((.((((..((((((.....)))))).....((((......)))).....-......)))).)))))........ ( -15.60) >consensus ACGCACC__UA__CUUUAUGCUCGGAUAAUUGCGAGCAUUUGUCUUGGGCGUACAGCCAAAUCCA_________AAUCCA_AACCAAAUUACGGAUUCGCAUUUC .............((.(((((((((((((.((....)).))))).)))))))).))................................................. (-12.27 = -12.33 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:09 2006