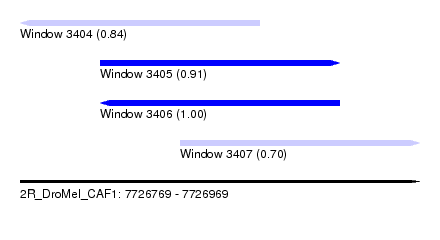
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,726,769 – 7,726,969 |
| Length | 200 |
| Max. P | 0.998941 |

| Location | 7,726,769 – 7,726,889 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -27.96 |
| Energy contribution | -27.84 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7726769 120 - 20766785 UUCAAUACACACCUGCGAUCGGGAUGGUCGACUUCGCGAGAAGAAACCAUUUUGUGCCAGUGAGUGUUGACAGUUUUCGAAAAUGUACACCCUUUCAAAUCGAACUAAGAAUAUGAAACA .(((((((.(((..(((..(((((((((...((((....))))..))))))))))))..))).)))))))..(((((.((((..(....)..))))...((.......))....))))). ( -30.60) >DroSec_CAF1 16805 120 - 1 UUCAAUACACACCUGCGAUCGGGAUGGUCGCCUUCGCGAGAAGAAACCAUUUUGUGCCAGUGAGUGUUGACAGUUUUCGAAAAUGUACACCCUUUACAAUAGAACUAGGAAUAUAAAACA .(((((((.(((..(((..(((((((((...((((....))))..))))))))))))..))).))))))).((((((......((((.......))))..)))))).............. ( -30.10) >DroSim_CAF1 15096 120 - 1 UUCAAUACACACCUGCGAUCGAGAUGGUCGCCUUCGCGAGAAGAAACCAUUUUGUGCCAGUGAGUGUUGACAGUUUUCGAAAAUGUACACCCUUUACAAUAGAACUAAGAAUAUAAAACA .(((((((.(((..(((..(((((((((...((((....))))..))))))))))))..))).))))))).((((((......((((.......))))..)))))).............. ( -31.00) >DroEre_CAF1 10321 97 - 1 CUCAAUACACACCUGCGAUCGGGAUGGUCGCCUUCGCGAGAAGAAGCCAUUUUGUGCCAGUGAGUGUUGACAGUUUUCG---------------UAAAUGUAUACUA--------AAACC .(((((((.(((..(((..(((((((((...((((....))))..))))))))))))..))).)))))))..(((((.(---------------((......))).)--------)))). ( -31.70) >DroYak_CAF1 10548 102 - 1 CUCAAUACACACCUGCGAUCGGGAUGGUCGCCUGCGCGAGAAGAAACCAUUUUGUGCCAGUGAGUGUUGACAGUGUACA----------CCCUUUAAGAUCGAACUA--------ACCUA .(((((((.(((..((((((.....))))))..((((((((.(....).))))))))..))).))))))).........----------..................--------..... ( -25.30) >consensus UUCAAUACACACCUGCGAUCGGGAUGGUCGCCUUCGCGAGAAGAAACCAUUUUGUGCCAGUGAGUGUUGACAGUUUUCGAAAAUGUACACCCUUUAAAAUAGAACUA_GAAUAU_AAACA .(((((((.(((..(((..(((((((((...((((....))))..))))))))))))..))).))))))).................................................. (-27.96 = -27.84 + -0.12)

| Location | 7,726,809 – 7,726,929 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -28.96 |
| Energy contribution | -29.96 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7726809 120 + 20766785 UCGAAAACUGUCAACACUCACUGGCACAAAAUGGUUUCUUCUCGCGAAGUCGACCAUCCCGAUCGCAGGUGUGUAUUGAAUUUCCCUGCAACACAAAUCCGCGAUUGCAUUGGAAAAUCA ..((......((((.((.((((.((.....((((((.((((....))))..)))))).......)).)))).)).)))).(((((.((((((.(......).).)))))..))))).)). ( -28.60) >DroSec_CAF1 16845 120 + 1 UCGAAAACUGUCAACACUCACUGGCACAAAAUGGUUUCUUCUCGCGAAGGCGACCAUCCCGAUCGCAGGUGUGUAUUGAAUUUCCCUGCAAUACAAAUCCGCGAUUGCAUUGGAAAAUCA ........(((((........))))).....((((((....((((....))))(((...(((((((.(((.(((((((..........))))))).))).)))))))...))).)))))) ( -34.20) >DroSim_CAF1 15136 120 + 1 UCGAAAACUGUCAACACUCACUGGCACAAAAUGGUUUCUUCUCGCGAAGGCGACCAUCUCGAUCGCAGGUGUGUAUUGAAUUUCCCUGCAAUACAAAUCCGCGAUUGCAUUGGAAAAUCA ........(((((........))))).....((((((....((((....))))(((...(((((((.(((.(((((((..........))))))).))).)))))))...))).)))))) ( -34.20) >DroEre_CAF1 10339 119 + 1 -CGAAAACUGUCAACACUCACUGGCACAAAAUGGCUUCUUCUCGCGAAGGCGACCAUCCCGAUCGCAGGUGUGUAUUGAGUUUCCUUGCAAUACAAAUCCGCGAUUGCAUUGGAAAAUCA -.......(((((........)))))...............((((....))))(((...(((((((.(((.(((((((..........))))))).))).)))))))...)))....... ( -32.30) >DroYak_CAF1 10571 119 + 1 -UGUACACUGUCAACACUCACUGGCACAAAAUGGUUUCUUCUCGCGCAGGCGACCAUCCCGAUCGCAGGUGUGUAUUGAGUUUCCUUGCAAGACAAAUCCCCGAUUGCAUUGGAAAAUCA -.((((..(((((........))))).................((((..((((.(.....).))))..))))))))(((.(((((.(((((.............)))))..))))).))) ( -26.42) >consensus UCGAAAACUGUCAACACUCACUGGCACAAAAUGGUUUCUUCUCGCGAAGGCGACCAUCCCGAUCGCAGGUGUGUAUUGAAUUUCCCUGCAAUACAAAUCCGCGAUUGCAUUGGAAAAUCA ........(((((........))))).....((((((....((((....))))(((...(((((((.(((.(((((((..........))))))).))).)))))))...))).)))))) (-28.96 = -29.96 + 1.00)

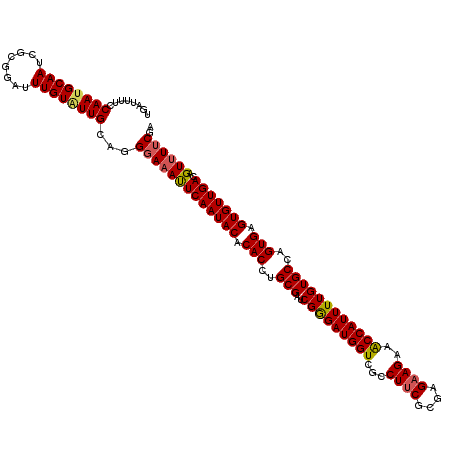
| Location | 7,726,809 – 7,726,929 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -38.48 |
| Consensus MFE | -37.32 |
| Energy contribution | -37.44 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7726809 120 - 20766785 UGAUUUUCCAAUGCAAUCGCGGAUUUGUGUUGCAGGGAAAUUCAAUACACACCUGCGAUCGGGAUGGUCGACUUCGCGAGAAGAAACCAUUUUGUGCCAGUGAGUGUUGACAGUUUUCGA ..(((((((..(((((.((((....))))))))))))))))(((((((.(((..(((..(((((((((...((((....))))..))))))))))))..))).))))))).......... ( -40.60) >DroSec_CAF1 16845 120 - 1 UGAUUUUCCAAUGCAAUCGCGGAUUUGUAUUGCAGGGAAAUUCAAUACACACCUGCGAUCGGGAUGGUCGCCUUCGCGAGAAGAAACCAUUUUGUGCCAGUGAGUGUUGACAGUUUUCGA ..(((((((..((((((.(((....))))))))))))))))(((((((.(((..(((..(((((((((...((((....))))..))))))))))))..))).))))))).......... ( -39.10) >DroSim_CAF1 15136 120 - 1 UGAUUUUCCAAUGCAAUCGCGGAUUUGUAUUGCAGGGAAAUUCAAUACACACCUGCGAUCGAGAUGGUCGCCUUCGCGAGAAGAAACCAUUUUGUGCCAGUGAGUGUUGACAGUUUUCGA ..(((((((..((((((.(((....))))))))))))))))(((((((.(((..(((..(((((((((...((((....))))..))))))))))))..))).))))))).......... ( -40.00) >DroEre_CAF1 10339 119 - 1 UGAUUUUCCAAUGCAAUCGCGGAUUUGUAUUGCAAGGAAACUCAAUACACACCUGCGAUCGGGAUGGUCGCCUUCGCGAGAAGAAGCCAUUUUGUGCCAGUGAGUGUUGACAGUUUUCG- ........((((((((........))))))))...(((((((((((((.(((..(((..(((((((((...((((....))))..))))))))))))..))).)))))))..)))))).- ( -40.60) >DroYak_CAF1 10571 119 - 1 UGAUUUUCCAAUGCAAUCGGGGAUUUGUCUUGCAAGGAAACUCAAUACACACCUGCGAUCGGGAUGGUCGCCUGCGCGAGAAGAAACCAUUUUGUGCCAGUGAGUGUUGACAGUGUACA- ....(((((..(((((.(((....)))..))))).))))).(((((((.(((..((((((.....))))))..((((((((.(....).))))))))..))).))))))).........- ( -32.10) >consensus UGAUUUUCCAAUGCAAUCGCGGAUUUGUAUUGCAGGGAAAUUCAAUACACACCUGCGAUCGGGAUGGUCGCCUUCGCGAGAAGAAACCAUUUUGUGCCAGUGAGUGUUGACAGUUUUCGA ........((((((((........))))))))...(((((((((((((.(((..(((..(((((((((...((((....))))..))))))))))))..))).)))))))..)))))).. (-37.32 = -37.44 + 0.12)


| Location | 7,726,849 – 7,726,969 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -27.86 |
| Energy contribution | -28.90 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7726849 120 + 20766785 CUCGCGAAGUCGACCAUCCCGAUCGCAGGUGUGUAUUGAAUUUCCCUGCAACACAAAUCCGCGAUUGCAUUGGAAAAUCACUAAAUUGUUCGAUCGUGUGAAGAAUCGUCCAUUUUCCAA .(((((..((((((((...(((((((.(((.(((.(((..........))).))).))).)))))))...)))......((......)))))))..)))))................... ( -28.10) >DroSec_CAF1 16885 120 + 1 CUCGCGAAGGCGACCAUCCCGAUCGCAGGUGUGUAUUGAAUUUCCCUGCAAUACAAAUCCGCGAUUGCAUUGGAAAAUCACUAAAUUGUUCGAUCGUGUGAAGAAUCGUCCAUUUGCCAA ...(((((((((((((...(((((((.(((.(((((((..........))))))).))).)))))))...)))....((((...(((....)))...))))....))).)).)))))... ( -35.40) >DroSim_CAF1 15176 120 + 1 CUCGCGAAGGCGACCAUCUCGAUCGCAGGUGUGUAUUGAAUUUCCCUGCAAUACAAAUCCGCGAUUGCAUUGGAAAAUCACUAAAUUGUUCGAUCGCGUAAAGAAUCGUCCAUUUUCCAA .((((....))))......(((((((.(((.(((((((..........))))))).))).)))))))..((((((((.........((..(((((.......).))))..)))))))))) ( -33.60) >DroEre_CAF1 10378 120 + 1 CUCGCGAAGGCGACCAUCCCGAUCGCAGGUGUGUAUUGAGUUUCCUUGCAAUACAAAUCCGCGAUUGCAUUGGAAAAUCACUAAAUUGUUCGAUCGAGUGAAGAACCGUCCAUUUUCCAA .((((....))))......(((((((.(((.(((((((..........))))))).))).)))))))..(((((((((((((..(((....)))..))))).((....))..)))))))) ( -36.00) >DroYak_CAF1 10610 120 + 1 CUCGCGCAGGCGACCAUCCCGAUCGCAGGUGUGUAUUGAGUUUCCUUGCAAGACAAAUCCCCGAUUGCAUUGGAAAAUCACUAAGUUGUUUGAUCGAGUGAAGAACCGUCCAUUUUCCAA ((((..((((((((((..((.......))..))...(((.(((((.(((((.............)))))..))))).)))....))))))))..)))).(((((........)))))... ( -28.72) >consensus CUCGCGAAGGCGACCAUCCCGAUCGCAGGUGUGUAUUGAAUUUCCCUGCAAUACAAAUCCGCGAUUGCAUUGGAAAAUCACUAAAUUGUUCGAUCGAGUGAAGAAUCGUCCAUUUUCCAA .((((....))))......(((((((.(((.(((((((..........))))))).))).)))))))..(((((((((......(((.((((......)))).))).....))))))))) (-27.86 = -28.90 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:07 2006