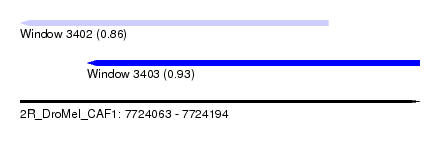
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,724,063 – 7,724,194 |
| Length | 131 |
| Max. P | 0.927657 |

| Location | 7,724,063 – 7,724,164 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -14.88 |
| Energy contribution | -14.43 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
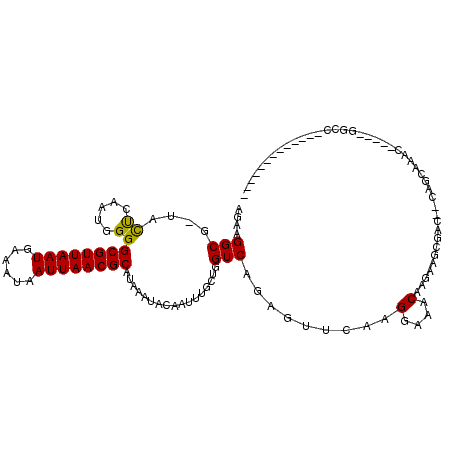
>2R_DroMel_CAF1 7724063 101 - 20766785 AGAAGGCGCUACUCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACAAUUUGCUGGUCAGAGUUCAAGGAAACAAGAAGCGAACACAGCAAAC-----GGCC------------- ....(((.............((((((((......))))))))..........(((((((((..(..(((..(....)..))).)..)).))))))).-----.)))------------- ( -27.00) >DroPse_CAF1 9787 116 - 1 GGAAGGCG-UAUCCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACGAUUCUCUGGUCAGAGUUCAAGGAAACAAGCAGCGAG--CAGCAAACAAGAAGGGCCAAAGGGCCAAAG .....(((-(.(((......((((((((......))))))))........((..((((....)))).))..))).))..((.....)--).))..........((((....)))).... ( -31.10) >DroEre_CAF1 7835 99 - 1 AGAAGGCGCCACUCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACAAUUUGCUAGUCAGAGUUCAAGGAAACAAGAAGCGAC--CAGCAAUC-----GGCC------------- .......(((..........((((((((......))))))))...........(((((.(((.(..(((..(....)..))).))))--.)))))..-----))).------------- ( -26.20) >DroYak_CAF1 7747 99 - 1 AGAAGGCGCUACUCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACAAUUUGCUGGUCAGAGUUCAAGGAAACAAGAAGCGAC--CAGCAAAC-----GGCC------------- ....(((.............((((((((......))))))))..........((((((((((.(..(((..(....)..))).))))--))))))).-----.)))------------- ( -32.40) >DroAna_CAF1 7834 97 - 1 AGAAGGCG-UACUCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACAAUUUGCUGGUCAGAGUUCAAGGAAACAAGGAC-GUC--GGACCAGC-----CGCU------------- ....((((-...........((((((((......)))))))).............(((((((.((((((..(....)..))))-.))--.)))))))-----))))------------- ( -34.00) >DroPer_CAF1 9304 116 - 1 GGAAGGCG-UAUCCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACGAUUCUCUGGUCAGAGUUCAAGGAAACAAGCAGCGAG--CAGCAAACAAGAAGGGCCAAAGGGCCAAAG .....(((-(.(((......((((((((......))))))))........((..((((....)))).))..))).))..((.....)--).))..........((((....)))).... ( -31.10) >consensus AGAAGGCG_UACUCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACAAUUUGCUGGUCAGAGUUCAAGGAAACAAGAAGCGAC__CAGCAAAC_____GGCC_____________ ....(((....((.....))((((((((......)))))))).................))).........(....).......................................... (-14.88 = -14.43 + -0.44)

| Location | 7,724,085 – 7,724,194 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -23.21 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.27 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
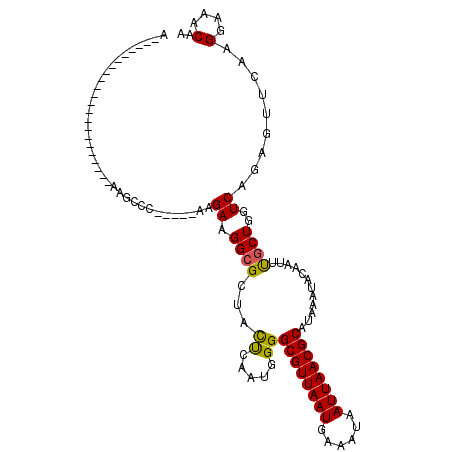
>2R_DroMel_CAF1 7724085 109 - 20766785 AACCAAAACCGUAGCCAAAGCCAAAGCCC-----AAGAAGGCGCUACUCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACAAUUUGCUGGUCAGAGUUCAAGGAAACAA .((((.....(((((....(((.......-----.....))))))))........((((((((......))))))))...............))))..........(....).. ( -23.30) >DroPse_CAF1 9825 85 - 1 -----------------------UGGCCA-----AGGAAGGCG-UAUCCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACGAUUCUCUGGUCAGAGUUCAAGGAAACAA -----------------------((((((-----..(((..((-(((((....))((((((((......))))))))....))))).)))..))))))........(....).. ( -29.10) >DroSim_CAF1 11948 103 - 1 AUCCAAAGCCG------AAGCCAAAGCCC-----AAGAAGGCGCUACUCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACAAUUUGCUGGUCAGAGUUCAAGGAAACAA .(((..((((.------..((((...(((-----(...((......))...))))((((((((......))))))))...............))))..).)))...)))..... ( -23.50) >DroEre_CAF1 7855 82 - 1 ---------------------------CC-----AAGAAGGCGCCACUCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACAAUUUGCUAGUCAGAGUUCAAGGAAACAA ---------------------------..-----..((.(((.(((.....))).((((((((......)))))))).............)))..)).........(....).. ( -16.80) >DroYak_CAF1 7767 98 - 1 ACCCAAGAC-----------ACGAUGCCC-----AAGAAGGCGCUACUCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACAAUUUGCUGGUCAGAGUUCAAGGAAACAA .((((....-----------....((((.-----.....))))........))))((((((((......)))))))).............................(....).. ( -21.39) >DroAna_CAF1 7853 93 - 1 GG--------G------------UAGCCCCUUUGGAGAAGGCG-UACUCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACAAUUUGCUGGUCAGAGUUCAAGGAAACAA ((--------.------------(((((((....(((......-..)))...)))((((((((......)))))))).............)))).)).........(....).. ( -25.20) >consensus A______________________AAGCCC_____AAGAAGGCGCUACUCAAUGGGGCGUUAAUGAAAUAAUUAACGCAUAAAUACAAUUUGCUGGUCAGAGUUCAAGGAAACAA ....................................((.((((...((.....))((((((((......))))))))............))))..)).........(....).. (-15.52 = -15.27 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:03 2006