| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,721,926 – 7,722,086 |
| Length | 160 |
| Max. P | 0.941110 |

| Location | 7,721,926 – 7,722,046 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.92 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
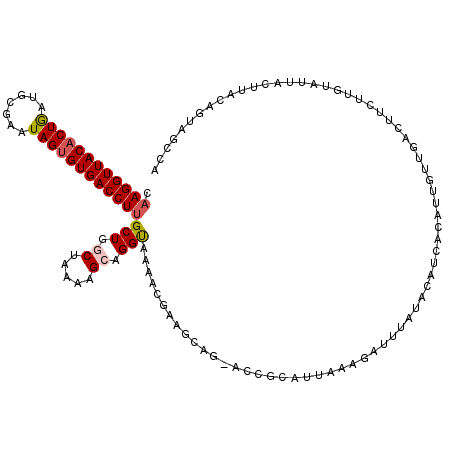
>2R_DroMel_CAF1 7721926 120 + 20766785 UGACAAUGAAAUGCACUUAUUUAUACCAUUUUUAAUUCGCCAAGGUUACACUGAUGCGAAUAGUGUGACCUUGCUGGCUAAAAGCAGGUUAAACGAAGCAGCACCGCAUUAAAGACUUAU ....((((...(((.(((......(((..........((.(((((((((((((.......))))))))))))).))((.....)).)))......)))..)))...)))).......... ( -29.20) >DroSec_CAF1 8250 114 + 1 -UACAAUGGCAUGCACUUAUUUAUACCUUUUUUAAUUUGCCAAGGUUACACUGAUGAGAAUAGUGUGACCUUGCUGGCGAAAAGCAGGCACAACGAA-----ACCGCAAUAAAGAUUUAU -.....((((.(((......(((.........)))((((((((((((((((((.......)))))))))))...)))))))..))).)).)).....-----.................. ( -26.70) >DroSim_CAF1 9814 114 + 1 -UGCAAUGGCAUGCAUUUAUUUAUACCUUUUUUAAUUCGCCAAGGUUACACUGAUGCGAAUAGUGUGACCUUGCUGGCGAAAAGCAGGCAAAACGAA-----ACCGCAUUAAAGAUUUAU -((((......)))).............(((((((((((((((((((((((((.......)))))))))))...)))))))..((.((.........-----.)))).)))))))..... ( -30.70) >DroEre_CAF1 5751 115 + 1 UGACAACGAAAUGCACUUAUUUAUACCUCAUUUAGUUCGCCAAGGUUACACUACUUCGAAUAGAGUGACCUUGCUAGCUAAAAGAAGGUAAAACGAAGCAG-----CAUUAAAGAUUUAU .........(((((.(((.....((((((.(((((((.((.((((((((.(((.......))).)))))))))).))))))).).))))).....)))..)-----)))).......... ( -31.60) >DroYak_CAF1 5637 117 + 1 ---CAACGAAAUGCACAUAUUUAUACCUUAUUUUAUUUGUCUAGGUUACACUAAUGCGACUAGUGUGACCUUGCUAACUAAAAGAAGGUAAAACGAAGCAGCACCGCAUUAAAGAUUUAU ---......(((((.........((((((.(((((.(((.(.(((((((((((.......))))))))))).).))).))))).))))))....(......)...))))).......... ( -28.40) >consensus _GACAAUGAAAUGCACUUAUUUAUACCUUUUUUAAUUCGCCAAGGUUACACUGAUGCGAAUAGUGUGACCUUGCUGGCUAAAAGCAGGUAAAACGAAGCAG_ACCGCAUUAAAGAUUUAU .......................((((((((((..((.(.(((((((((((((.......)))))))))))))).))..))))).))))).............................. (-19.12 = -19.92 + 0.80)


| Location | 7,721,926 – 7,722,046 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -15.93 |
| Energy contribution | -15.89 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7721926 120 - 20766785 AUAAGUCUUUAAUGCGGUGCUGCUUCGUUUAACCUGCUUUUAGCCAGCAAGGUCACACUAUUCGCAUCAGUGUAACCUUGGCGAAUUAAAAAUGGUAUAAAUAAGUGCAUUUCAUUGUCA .........(((((..((((.((((.((((((((...((((((...(((((((.(((((.........))))).))))).))...))))))..))).)))))))))))))..)))))... ( -31.30) >DroSec_CAF1 8250 114 - 1 AUAAAUCUUUAUUGCGGU-----UUCGUUGUGCCUGCUUUUCGCCAGCAAGGUCACACUAUUCUCAUCAGUGUAACCUUGGCAAAUUAAAAAAGGUAUAAAUAAGUGCAUGCCAUUGUA- ............((((.(-----(...((((((((..(((..((((...((((.(((((.........))))).)))))))).....)))..))))))))..)).))))..........- ( -28.30) >DroSim_CAF1 9814 114 - 1 AUAAAUCUUUAAUGCGGU-----UUCGUUUUGCCUGCUUUUCGCCAGCAAGGUCACACUAUUCGCAUCAGUGUAACCUUGGCGAAUUAAAAAAGGUAUAAAUAAAUGCAUGCCAUUGCA- ...........(((((..-----....((.(((((....(((((((...((((.(((((.........))))).))))))))))).......))))).)).....)))))((....)).- ( -29.80) >DroEre_CAF1 5751 115 - 1 AUAAAUCUUUAAUG-----CUGCUUCGUUUUACCUUCUUUUAGCUAGCAAGGUCACUCUAUUCGAAGUAGUGUAACCUUGGCGAACUAAAUGAGGUAUAAAUAAGUGCAUUUCGUUGUCA ..........((((-----(.((((.((((((((((..(((((...(((((((.((.((((.....)))).)).))))).))...))))).)))))).)))))))))))))......... ( -30.40) >DroYak_CAF1 5637 117 - 1 AUAAAUCUUUAAUGCGGUGCUGCUUCGUUUUACCUUCUUUUAGUUAGCAAGGUCACACUAGUCGCAUUAGUGUAACCUAGACAAAUAAAAUAAGGUAUAAAUAUGUGCAUUUCGUUG--- .........(((((..((((.((....((.((((((.(((((.....(.((((.((((((((...)))))))).)))).).....))))).)))))).))....))))))..)))))--- ( -31.10) >consensus AUAAAUCUUUAAUGCGGU_CUGCUUCGUUUUACCUGCUUUUAGCCAGCAAGGUCACACUAUUCGCAUCAGUGUAACCUUGGCGAAUUAAAAAAGGUAUAAAUAAGUGCAUUUCAUUGUA_ .........(((((.............((.(((((..(((..((((...((((.(((((.........))))).)))))))).....)))..))))).))............)))))... (-15.93 = -15.89 + -0.04)

| Location | 7,721,966 – 7,722,086 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.64 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.941110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7721966 120 + 20766785 CAAGGUUACACUGAUGCGAAUAGUGUGACCUUGCUGGCUAAAAGCAGGUUAAACGAAGCAGCACCGCAUUAAAGACUUAUUCAACACAUGGUUGACUUCUUGUAUUACUUACAGUAGCCA (((((((((((((.......)))))))))))))..(((((...(.((((...((((.((......)).............(((((.....)))))....))))...)))).)..))))). ( -33.50) >DroSec_CAF1 8289 115 + 1 CAAGGUUACACUGAUGAGAAUAGUGUGACCUUGCUGGCGAAAAGCAGGCACAACGAA-----ACCGCAAUAAAGAUUUAUACAUCACAUUGUUGACUUCUUGUAUUACUUACAGUAGUCA .((((((((((((.......))))))))))))(((.((.....)).)))........-----...(((((...(((......)))..)))))(((((..(((((.....))))).))))) ( -29.60) >DroSim_CAF1 9853 115 + 1 CAAGGUUACACUGAUGCGAAUAGUGUGACCUUGCUGGCGAAAAGCAGGCAAAACGAA-----ACCGCAUUAAAGAUUUAUACAUCACAUUGUUGACUUCUUGUAUUACUUACAGUAGUCA .((((((((((((.......))))))))))))(((.((.....)).)))........-----...........(((......))).......(((((..(((((.....))))).))))) ( -28.00) >DroEre_CAF1 5791 93 + 1 CAAGGUUACACUACUUCGAAUAGAGUGACCUUGCUAGCUAAAAGAAGGUAAAACGAAGCAG-----CAUUAAAGAUUUAUACAU----------------------UCUUUCAGCAGCCA .((((((((.(((.......))).))))))))(((.(((.((((((.(((((.(.((....-----..))...).)))))...)----------------------))))).)))))).. ( -23.20) >DroYak_CAF1 5674 120 + 1 CUAGGUUACACUAAUGCGACUAGUGUGACCUUGCUAACUAAAAGAAGGUAAAACGAAGCAGCACCGCAUUAAAGAUUUAUACAUCACAUUGUUGAGUUCUUGUAUUACUUUCAGUAGCCA ..(((((((((((.......))))))))))).((((.((....((((((((.((((.((......))......((((((.(((......))))))))).)))).)))))))))))))).. ( -31.60) >consensus CAAGGUUACACUGAUGCGAAUAGUGUGACCUUGCUGGCUAAAAGCAGGUAAAACGAAGCAG_ACCGCAUUAAAGAUUUAUACAUCACAUUGUUGACUUCUUGUAUUACUUACAGUAGCCA .((((((((((((.......))))))))))))(((.((.....)).)))....................................................................... (-16.32 = -16.64 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:00 2006