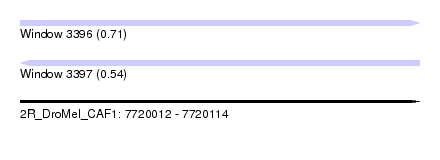
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,720,012 – 7,720,114 |
| Length | 102 |
| Max. P | 0.712491 |

| Location | 7,720,012 – 7,720,114 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -17.18 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7720012 102 + 20766785 AGGCAGAUUGACAGAUUGUGCGGAAAUUAGCUAAAG-AAUGCAAAGUCAAUUUCCGCUUUUGUCAAUUAUCCAAAGAACUUAAUA-GGCAUAAUAAUGUUAUUG .((..((((((((((....(((((((((..((....-.......))..))))))))).))))))))))..)).............-(((((....))))).... ( -28.00) >DroPse_CAF1 6201 103 + 1 UGGAAAACUGACAGCUUGAACGGAAAUUAGCUUAGGAAACGUAAAGUCAAUUUCCGGC-UUGUCAAUUAUACAACAGCCUUAUUUCGGAUUGAUUAUGCUAAUG (((.....(((((((.....((((((((..(((.(....)...)))..))))))))))-.))))).........(((((.......)).)))......)))... ( -22.20) >DroSim_CAF1 7526 102 + 1 AGGCAAAGUGACAGAUUGUGCGGAAAUUAGCUAAAG-AAUGCAAAGUCAAUUUCCGCUUUUGUCAAUUAUCCAAAGAACUUAAUA-GGCAUAAUAAUGUUAUUG .((.....(((((((....(((((((((..((....-.......))..))))))))).))))))).....)).............-(((((....))))).... ( -25.10) >DroEre_CAF1 3887 102 + 1 CGGCAAAGUGACAGAUUGUGCGGAAAUUAUCCAAAG-AAUGCAAAGUCAAUUUCCGCUUUUGUCAAAUAUCCAAAGAACUUUAUU-GGCAUAAUAAUGUUAUUG .((.....(((((((....(((((((((..(.....-........)..))))))))).))))))).....))............(-(((((....))))))... ( -22.12) >DroYak_CAF1 3690 102 + 1 CGGCAAAGUGACAGAUUGUGCGGAAAUUAGCCAGAG-AAUGCAAAGUCAAUUUCCGCUUUUGUCAAUUAUCCUAAGAACUUUAUU-GGCAUAAUAAUGUUAUUG .((((...(((((((....(((((((((..(.....-........)..))))))))).)))))))(((((.((((........))-)).)))))..)))).... ( -24.02) >DroPer_CAF1 5711 103 + 1 UGGAAAACUGACAGCUUGAACGGAAAUUAGCUUAGGAAACGUAAAGUCAAUUUCCGGC-UUGUCAAUUAUACAACAGCCUUAUUUCGGAUUGAUUAUGCUAAUG (((.....(((((((.....((((((((..(((.(....)...)))..))))))))))-.))))).........(((((.......)).)))......)))... ( -22.20) >consensus AGGCAAAGUGACAGAUUGUGCGGAAAUUAGCUAAAG_AAUGCAAAGUCAAUUUCCGCUUUUGUCAAUUAUCCAAAGAACUUAAUU_GGCAUAAUAAUGUUAUUG .((.....(((((((....(((((((((..((..(....)....))..))))))))).))))))).....))................................ (-17.18 = -18.07 + 0.89)

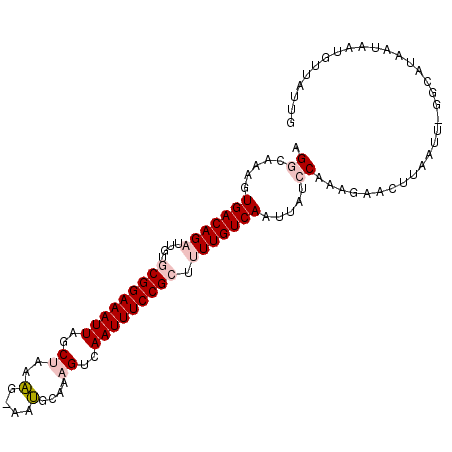
| Location | 7,720,012 – 7,720,114 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -19.92 |
| Consensus MFE | -14.92 |
| Energy contribution | -15.37 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7720012 102 - 20766785 CAAUAACAUUAUUAUGCC-UAUUAAGUUCUUUGGAUAAUUGACAAAAGCGGAAAUUGACUUUGCAUU-CUUUAGCUAAUUUCCGCACAAUCUGUCAAUCUGCCU ..................-.............((...(((((((...((((((((((.((.......-....)).))))))))))......)))))))...)). ( -20.60) >DroPse_CAF1 6201 103 - 1 CAUUAGCAUAAUCAAUCCGAAAUAAGGCUGUUGUAUAAUUGACAA-GCCGGAAAUUGACUUUACGUUUCCUAAGCUAAUUUCCGUUCAAGCUGUCAGUUUUCCA ((.((((....((.....))......)))).))...((((((((.-(((((((((((.(((..........))).))))))))).....))))))))))..... ( -21.00) >DroSim_CAF1 7526 102 - 1 CAAUAACAUUAUUAUGCC-UAUUAAGUUCUUUGGAUAAUUGACAAAAGCGGAAAUUGACUUUGCAUU-CUUUAGCUAAUUUCCGCACAAUCUGUCACUUUGCCU ..................-.............((.....(((((...((((((((((.((.......-....)).))))))))))......))))).....)). ( -18.10) >DroEre_CAF1 3887 102 - 1 CAAUAACAUUAUUAUGCC-AAUAAAGUUCUUUGGAUAUUUGACAAAAGCGGAAAUUGACUUUGCAUU-CUUUGGAUAAUUUCCGCACAAUCUGUCACUUUGCCG ....(((.(((((.....-))))).)))....((.....(((((...((((((((((.((.......-....)).))))))))))......))))).....)). ( -19.30) >DroYak_CAF1 3690 102 - 1 CAAUAACAUUAUUAUGCC-AAUAAAGUUCUUAGGAUAAUUGACAAAAGCGGAAAUUGACUUUGCAUU-CUCUGGCUAAUUUCCGCACAAUCUGUCACUUUGCCG ....(((.(((((.....-))))).)))....((.....(((((...((((((((((.((.......-....)).))))))))))......))))).....)). ( -19.50) >DroPer_CAF1 5711 103 - 1 CAUUAGCAUAAUCAAUCCGAAAUAAGGCUGUUGUAUAAUUGACAA-GCCGGAAAUUGACUUUACGUUUCCUAAGCUAAUUUCCGUUCAAGCUGUCAGUUUUCCA ((.((((....((.....))......)))).))...((((((((.-(((((((((((.(((..........))).))))))))).....))))))))))..... ( -21.00) >consensus CAAUAACAUUAUUAUGCC_AAUUAAGUUCUUUGGAUAAUUGACAAAAGCGGAAAUUGACUUUGCAUU_CUUUAGCUAAUUUCCGCACAAUCUGUCACUUUGCCA ................................((.....(((((...((((((((((.((............)).))))))))))......))))).....)). (-14.92 = -15.37 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:57 2006