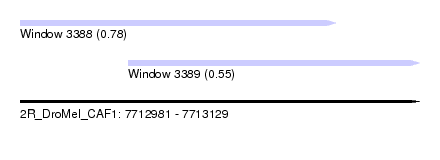
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,712,981 – 7,713,129 |
| Length | 148 |
| Max. P | 0.777784 |

| Location | 7,712,981 – 7,713,098 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -21.50 |
| Energy contribution | -21.72 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7712981 117 + 20766785 GCAGCUGGUGAGGAAAACACUGGUCACAUUCUGCACGAUCUGCACUUUGGACUCAGCAUUGUCCAGCAGCAUGGCCAAUAGCAGGUGCUGUUGGCCAUCCAGUUUCACGCUUGAGGA ((.((((.((.((......((((((.((...((((.....))))...))))).))).....)))).))))((((((((((((....))))))))))))..........))....... ( -42.50) >DroVir_CAF1 3475 117 + 1 GCAGCUGGUCAGGAAGACGCUGGUUACAUUCUGCACUAUUUGGAUCUUGGACUCAGCAUUGUCCAGCAACAUGGCCAGUAACAAAUGCUGUUGUCCGUCCAGCUGCACAUGUGAUGG ((((((((...(((....((((((..(((..(((.((....))....(((((........))))))))..)))))))))((((.....)))).)))..))))))))........... ( -40.40) >DroGri_CAF1 3247 117 + 1 GCAGCUGGUGAGGAAAACGCUCGUCACAUUCUGCACAAUUUGUAUCUUGGACUCGGCAUUAUCCAGCAGCAUCGCGAGCAACAGAUGCUGCUGUCCAUCCAGCUGCACUUCACACGA ((((((((...(((....(((.(((.((...((((.....))))...)))))..)))......((((((((((..........)))))))))))))..))))))))........... ( -46.10) >DroWil_CAF1 3135 117 + 1 GCAACUUGUUAGAAAAACACUCGUUACAUUUUGGACAAUUUGAAUUUUUGACUCUGCAUUGUCCAACAGCAUGGCCAGCAAUAGAUGUUGCUGCCCAUCCAGUUGUACCUCCCACGA (((((((((..((.......))...)))..(((((((((..((.........))...)))))))))..(.((((.(((((((....))))))).)))).)))))))........... ( -28.50) >DroMoj_CAF1 3284 117 + 1 GCAGCUGGUGAGGAAGACGCUCGUUACGUUCUGGACAAUUUGUAUCUUGGACUCGGCAUUGUCGAGUAGCAUGGCCAGUAACAGAUGCUGUUGUCCAUCCAACUGCACCUCCGAUGG ((((.((((((.((......)).))))....((((((((..((((((...(((.(((..(((......)))..))))))...)))))).))))))))..)).)))).((......)) ( -38.70) >DroAna_CAF1 2993 117 + 1 ACAGCUGGUGAGAAACACGCUUGUGACAUUUUGGACUAUUUGGAUCUUGGACUCGGCAUUGUCCAGGAGCAUGGCCAAAAGCAGAUGCUGCUGUCCGUCCAACUUAACAGCGGACGG (((((..(((.....)))((.(.((...((((((.((((.....((((((((........))))))))..)))))))))).)).).)).)))))((((((...........)))))) ( -40.60) >consensus GCAGCUGGUGAGGAAAACGCUCGUUACAUUCUGCACAAUUUGGAUCUUGGACUCGGCAUUGUCCAGCAGCAUGGCCAGUAACAGAUGCUGCUGUCCAUCCAGCUGCACAUCCGACGA ((((((((...(((((((....)))...))))...............(((((.(((((((........((...))........)))))))..))))).))))))))........... (-21.50 = -21.72 + 0.23)
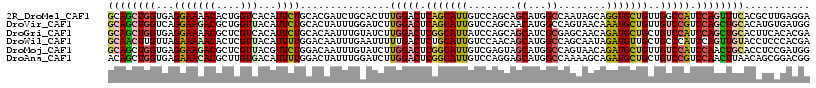
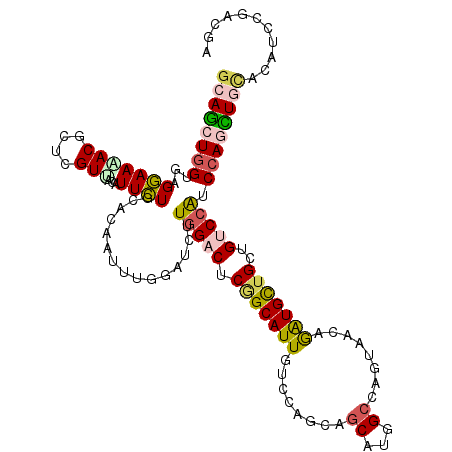
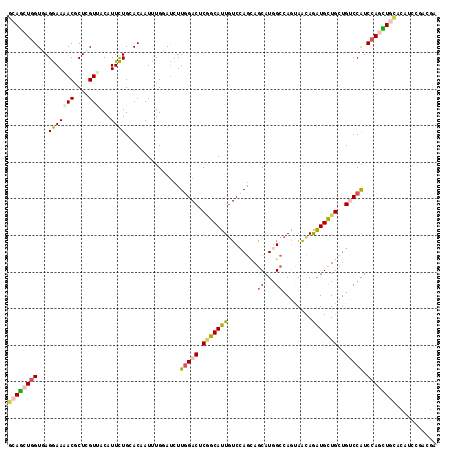
| Location | 7,713,021 – 7,713,129 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -12.97 |
| Energy contribution | -13.59 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7713021 108 + 20766785 UGCACUUUGGACUCAGCAUUGUCCAGCAGCAUGGCCAAUAGCAGGUGCUGUUGGCCAUCCAGUUUCACGCUUGAGGAAUCGACAUUCUUAUCCACCUCCAUUGCAUCG ((((...((((((((((..((.....))(.((((((((((((....)))))))))))).)........)).))))((((....)))).........)))).))))... ( -35.50) >DroVir_CAF1 3515 108 + 1 UGGAUCUUGGACUCAGCAUUGUCCAGCAACAUGGCCAGUAACAAAUGCUGUUGUCCGUCCAGCUGCACAUGUGAUGGAUCGCCAUCCGUGUCCACUUCCAUGGCAUCC (((((...((((.((((((((((((......)))......)).)))))))..)))))))))..(((.((((..(((((.(((.....))))))).)..)))))))... ( -35.00) >DroGri_CAF1 3287 105 + 1 UGUAUCUUGGACUCGGCAUUAUCCAGCAGCAUCGCGAGCAACAGAUGCUGCUGUCCAUCCAGCUGCACUUCACACGACUCUCCAC---UGUCCACGUCCAUGGCAUCC (((....(((((.((((......((((((((((..........))))))))))........))))..........(((.......---.)))...)))))..)))... ( -31.14) >DroYak_CAF1 3047 108 + 1 UGCACCUUGGACUCUGCAUUGUCCAGCAGCAUGGCCAAUAGCAGGUGCUGCUGGCCAUCCAACUUCACGCAUGAGGAAUCGAUGUCUUCAUCAACCUCCAUUGCAUCG .((...((((((........))))))..))(((((((.((((....)))).)))))))..........(((.((((....((((....))))..))))...))).... ( -36.40) >DroMoj_CAF1 3324 108 + 1 UGUAUCUUGGACUCGGCAUUGUCGAGUAGCAUGGCCAGUAACAGAUGCUGUUGUCCAUCCAACUGCACCUCCGAUGGAUCGCCUUGCUGGUCCACAUCCAUGGCAUCU (((....((((((((((...)))))))....((((((((((..(((((.((((......)))).)).((......)))))...))))))).)))...)))..)))... ( -33.90) >DroAna_CAF1 3033 108 + 1 UGGAUCUUGGACUCGGCAUUGUCCAGGAGCAUGGCCAAAAGCAGAUGCUGCUGUCCGUCCAACUUAACAGCGGACGGAUCGGUAUCCUCAUCUAUCUCCAUCGCAUCA ....((((((((........))))))))((((((.........(((((((..((((((((...........)))))))))))))))...........)))).)).... ( -37.45) >consensus UGCAUCUUGGACUCGGCAUUGUCCAGCAGCAUGGCCAAUAACAGAUGCUGCUGUCCAUCCAACUGCACAUCUGACGAAUCGCCAUCCUUAUCCACCUCCAUGGCAUCC (((...((((((........))))))..)))............(((((.((((......)))).........((((..................))))....))))). (-12.97 = -13.59 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:49 2006