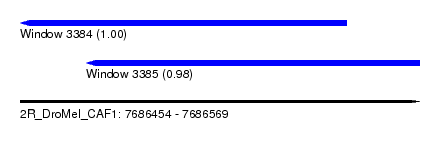
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,686,454 – 7,686,569 |
| Length | 115 |
| Max. P | 0.999372 |

| Location | 7,686,454 – 7,686,548 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 73.21 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -9.12 |
| Energy contribution | -9.08 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.44 |
| SVM decision value | 3.55 |
| SVM RNA-class probability | 0.999372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
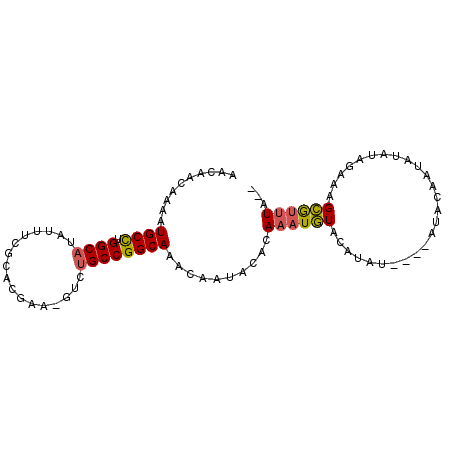
>2R_DroMel_CAF1 7686454 94 - 20766785 AACAACAAAAUGCCUGGCAUAUUUCGCACGUA-GUCUGCCGGCAAACAAUACACAAAUGUACAUAUGUAUGUACAAUGUGUGGAAAGCAUUUAUA .......((((((((((((.(((.(....).)-)).)))))).......((((((..(((((((....))))))).))))))....))))))... ( -26.10) >DroSec_CAF1 5417 88 - 1 AACAACAAAAUGCCUGGCAUAUUUCGCACGUA-GUCUGCCGGCAAACAAUACACAAAUGUACAUAU----GUGCAAUAUGUGGAAAGCGUUUA-- .......((((((((((((.(((.(....).)-)).)))))).........((((..(((((....----)))))...))))....)))))).-- ( -22.00) >DroEre_CAF1 7918 92 - 1 AAUAACAAAAUGCCUGGCAUAUUUCACACAAA-GUCUGCCGGCAAACAAUACACAAAUGUAUGUAGAUACAUACGAUAUACAGAAAGCGUUUA-- .......((((((((((((.((((......))-)).))))))...............(((((((....)))))))...........)))))).-- ( -19.70) >DroYak_CAF1 7893 89 - 1 AACAACAAAAUGCCUGGCAUAUUUCGCACGAA-GUCUGCCGGCAAACAAUACACAAAUGUACAUAC---GACACGAUAUAUAGGAGGCGUUUA-- .......((((((((((((.(((((....)))-)).)))).........((((....)))).....---...............)))))))).-- ( -20.80) >DroAna_CAF1 7798 82 - 1 AACAACAAAAUGCUUGGCAUUUUUCACGUCCACCCCAGCCGGCAAAAAC---CAAAAUAUCUAUA------CACCA--CCCCAGUGGUGCGUA-- ..........((((.(((...................))))))).....---.........((..------(((((--(....))))))..))-- ( -15.51) >consensus AACAACAAAAUGCCUGGCAUAUUUCGCACGAA_GUCUGCCGGCAAACAAUACACAAAUGUACAUAU____AUACAAUAUAUAGAAAGCGUUUA__ ..........((((.((((.................))))))))..........((((((..........................))))))... ( -9.12 = -9.08 + -0.04)

| Location | 7,686,473 – 7,686,569 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.25 |
| Mean single sequence MFE | -20.13 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.17 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
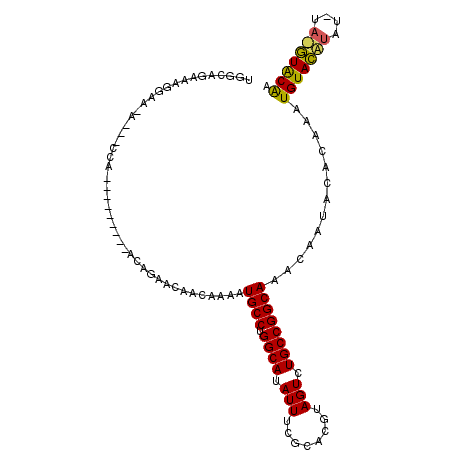
>2R_DroMel_CAF1 7686473 96 - 20766785 UGGCAGAAAGGAA-A---CCA---------ACAUAACAACAAAAUGCCUGGCAUAUUUCGCACGUAGUCUGCCGGCAAACAAUACACAAAUGUACAUAUGUAUGUACAA .((((((..(...-.---)..---------..............(((...((.......))..))).)))))).................(((((((....))))))). ( -20.10) >DroSec_CAF1 5434 92 - 1 UGGCAGAAAGGAA-A---CGA---------ACAGAACAACAAAAUGCCUGGCAUAUUUCGCACGUAGUCUGCCGGCAAACAAUACACAAAUGUACAUAU----GUGCAA .((((((..(...-.---)..---------..............(((...((.......))..))).)))))).................(((((....----))))). ( -19.10) >DroEre_CAF1 7935 109 - 1 UGGCAGAAAGGAAAAACACCAACACAGCAAGCAGAAUAACAAAAUGCCUGGCAUAUUUCACACAAAGUCUGCCGGCAAACAAUACACAAAUGUAUGUAGAUACAUACGA .((((((..((.......))......((..(((...........)))...))...............)))))).................(((((((....))))))). ( -21.20) >consensus UGGCAGAAAGGAA_A___CCA_________ACAGAACAACAAAAUGCCUGGCAUAUUUCGCACGUAGUCUGCCGGCAAACAAUACACAAAUGUACAUAU_UA_GUACAA ............................................((((.((((.(((........))).)))))))).............(((((((....))))))). (-17.60 = -17.17 + -0.44)

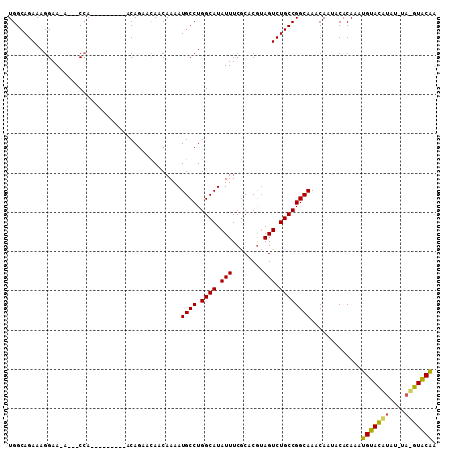
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:45 2006