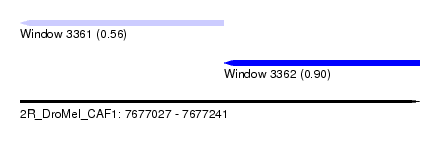
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,677,027 – 7,677,241 |
| Length | 214 |
| Max. P | 0.901647 |

| Location | 7,677,027 – 7,677,136 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.41 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.21 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.41 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7677027 109 - 20766785 GCUCCCAGG-UUACUGCCAAAUUUGCGAUAAA------ACGGGAGCUGCAAUGA-AUAUGUUCGUGGCGAUUGUUAAGAAUUAAUCGUGACUGCUGAUAGUUGCAUACAUAUAAACU--- ((((((...-(((.(((.......))).))).------..))))))((((((..-....((..((.(((((((........))))))).)).)).....))))))............--- ( -30.80) >DroPse_CAF1 5616 108 - 1 ACUCCCAGC-UUACUACUAAUUUAGCGAUUA-------AAGGGAGUUGCAAUGA-AUAUGUUCGUGAUGAUUGAACAGCAUAAAUCGUUGCUGCUGAUACUUGCAUACACUGGAAAA--- ((((((.((-(............))).....-------..)))))).(((((((-...((((((.......)))))).......)))))))(((........)))............--- ( -22.60) >DroGri_CAF1 7732 118 - 1 ACGCCCAAC-UUACUACCAAUUUAGCGAUUGUUUCCUUUUGGGAAGUGCAAUGU-GAAUGUUCGUUACGAUUUUCUAGACUAAAUCGCGACUGCUGAUAGUUGCAUACAAGCAUAGUUCU .(((...((-((....((((...((.((.....)))).)))).)))).....))-).(((((.((...(((((........)))))(((((((....)))))))..)).)))))...... ( -22.60) >DroWil_CAF1 6652 110 - 1 ACUCCCAAGUUUACUACCAAUUUAGCGAUAAUGG---UGAGGGAGAUGCAAUGAGAAAUGUUCGUUAUGAUUGCAAAGACUAAGUCAUAACUGCUGAUAGUUGCAGACA----AACU--- .(((((.((....))((((..(((....))))))---)..))))).((((((.......((..(((((((((..........))))))))).)).....))))))....----....--- ( -25.60) >DroMoj_CAF1 7924 116 - 1 ACGCCCAAU-UUACUACCAAUUUAGCGAUUGUCUUCGAAUGGGCGGUGCAAUGU-AAAUGUUCGUUAUGAUUUUGUAGACUAAAUUUUAACUGCUGAUAGUUGCAUACAAACAUAAAC-- .((((((..-...(((......)))(((......)))..))))))(((((((..-....((..((((.(((((........))))).)))).)).....)))))))............-- ( -25.20) >DroPer_CAF1 5617 108 - 1 ACUCCCAGC-UUACUACUAAUUUAGCGAUUA-------AAGGGAGUUGCAAUGA-AUAUGUUCGUGAUGAUUGAACAGCAUAAAUCGUUGCUGCUGAUACUUGCAUACACUGGAACA--- ((((((.((-(............))).....-------..)))))).(((((((-...((((((.......)))))).......)))))))(((........)))............--- ( -22.60) >consensus ACUCCCAAC_UUACUACCAAUUUAGCGAUUAU______AAGGGAGGUGCAAUGA_AAAUGUUCGUGAUGAUUGUAAAGAAUAAAUCGUAACUGCUGAUAGUUGCAUACAAACAAACA___ .(((((..................................))))).((((((.......((..(((((((((..........))))))))).)).....))))))............... (-17.54 = -17.21 + -0.33)

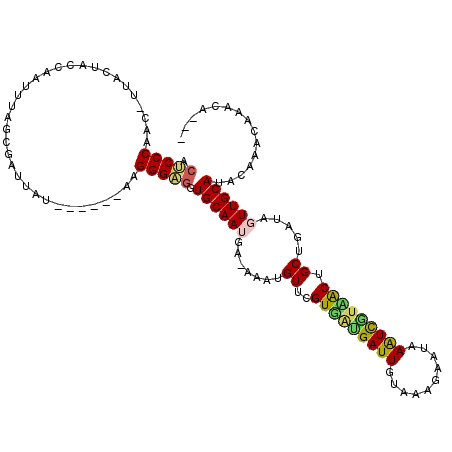
| Location | 7,677,136 – 7,677,241 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -19.98 |
| Energy contribution | -19.32 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7677136 105 - 20766785 AUUCUU-CAAAAAUUACCAUUCUAAAAUCUUAUUUACGUGGUUAGAAACGUUUCCAAGUGAUCAGCCUCUCUGGAAAUGCUUCAUGCCGCGUUAGAAAAACGACGA ......-............(((((...........(((((((..(((.((((((((.(.((......)).))))))))).)))..))))))))))))......... ( -23.90) >DroVir_CAF1 6898 104 - 1 AUCUUUACGAUUGUAUAUGUAGUUAGAU-AUAUUUACAUGGC-CGUAGAGUUCCCAGUUGUACGCAUAUACUGGUAACGCUUCAUGCCAUGUUAGAUCAAACAAUA .....................(((.(((-......(((((((-...((.(((.(((((((....))...))))).))).))....)))))))...))).))).... ( -23.80) >DroSec_CAF1 6852 105 - 1 AUUCUU-CGAAAAUUACCAUUCUAAAAUCUUAUUUACGUGGUUAGAAACGUUUCCAAGUGAACAGCCUCUCUGGAAAUGCUUCAUGCCGCGUUAGAAAAACGACGA ..((.(-((....(((......)))..(((.....(((((((..(((.((((((((.(.((......)).))))))))).)))..))))))).)))....))).)) ( -24.30) >DroSim_CAF1 6540 105 - 1 AUUAUU-CAAAAAUUACCAUUCUAAAAUCUUAUUUACGUGGUUAGAAACGUUUCCAAGUGAACAGCCUCUCUGGAAAUGCUUCAUGCCGCGUUAGAAAAACGACGA ......-............(((((...........(((((((..(((.((((((((.(.((......)).))))))))).)))..))))))))))))......... ( -24.10) >DroEre_CAF1 6856 105 - 1 AUUCCU-CCAAUAUGAUUAUUCUAACAUCUUAUUUACGUGGUCAAGAACGUUUCCAAGUGAACAGCCUCUCUGGAAAUGCUUCAUGCCGCGUUAGAAAAACGACGA ......-............(((((...........((((((.((.(((((((((((.(.((......)).))))))))).))).)))))))))))))......... ( -23.90) >DroYak_CAF1 6843 105 - 1 AUUCCU-CCAAAAUUAUUAUUCUAAAAUACUAUUUACGUGGUCAGAAACGUUUCUAAGUGAACAGCCUCUCUGGAAAUGCUUCAUGCCGCGUCAGAAAAACGACGA ......-.........................((((((((((..(((.((((((((.(.((......)).))))))))).)))..))))))).))).......... ( -20.40) >consensus AUUCUU_CAAAAAUUACCAUUCUAAAAUCUUAUUUACGUGGUUAGAAACGUUUCCAAGUGAACAGCCUCUCUGGAAAUGCUUCAUGCCGCGUUAGAAAAACGACGA ................................((((((((((..(((.(((((((.((.((......)).))))))))).)))..))))))).))).......... (-19.98 = -19.32 + -0.66)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:22 2006