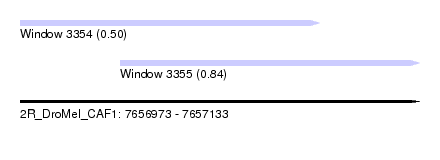
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,656,973 – 7,657,133 |
| Length | 160 |
| Max. P | 0.839184 |

| Location | 7,656,973 – 7,657,093 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -34.99 |
| Consensus MFE | -20.38 |
| Energy contribution | -21.66 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7656973 120 + 20766785 CGUGAUGCGCGCUCUUAUUGACUUCAAGUACAUUGAAAAGAUAUUCCUGAAUGUAACGCUUCGAGAGGACAAGUGCAACAAGCGGAGCGUGGAGUUCCGUAUGCUGGGCAACGAGCAGUU ..((.(((((..((((.((((.(((((.....)))))...(((((....)))))......)))).))))...))))).)).(((((((.....))))))).((((.(....).))))... ( -35.20) >DroVir_CAF1 117 120 + 1 CGUUGAACAUGCAUUGAUUGACUUCAAGUACAUGGAGAAGAUAUUCCUAAAUGUAACGCUACGCGAGAACAAAUGCAACAAGCGCAGCGUCGAAUUCCGCAUGCUGGGCAACGAGCAGUU (((((..((.((((.(...((.(((...(((((((((......))))...)))))(((((.(((.................))).))))).))).))..)))))))..)))))....... ( -30.13) >DroGri_CAF1 117 120 + 1 UGUGGAGCACGCCCUGAUCGACUUCAAAUACAUGGAAAAGAUAUUCUUAACUGUGUCGCUGCGCGAGAAUAAAUGCAAUAAGCGCAGCGUCGAAUUUCGCAUGCUUGGCAACGAGCAAUU (((((((((.....)).(((((.......((((((..(((.....)))..)))))).(((((((.................)))))))))))).)))))))((((((....))))))... ( -38.93) >DroWil_CAF1 1363 120 + 1 UGUGGAACAUGCCCUGAUUGACUUUAAAUACAUGGAGAAGAUCUAUUUAAAUGUGGUCAUGCGGGAGGAUAAAUCGAAUAAACGAAGCGUUGAGUUCCGUAUGCUUGGCAAUGAGCAGUU ...(((((((((((((..(((((.....((.(((((.....))))).)).....)))))..))))........(((......))).))))...)))))...(((((......)))))... ( -29.30) >DroMoj_CAF1 117 120 + 1 CGUGGAGCAUGCAUUGAUUGACUUCAAGUACAUGGAAAAAAUAUUCCUAAAUGUAACGUUGCGCGAAAGUAAAUGCAACAAGCGCAGUGUCGAAUUUCGCAUGCUUGGCAACGAGCAGUU .((.((((((((.....(((((......(((((((((......))))...)))))..(((((((....))....))))).........))))).....)))))))).))........... ( -34.70) >DroAna_CAF1 117 120 + 1 UGUGGAGCAUGCGCUAAUUGACUUCAAGUACAUGGAAAAGAUCUUCCUGAACGUAACGCUGCGGGAGGACAAGUGCAAUAAGCGCAGUGUGGAGUUCAGGAUGCUUGGCAACGAGCAAUU .......((((..((...((....))))..))))..........((((((((...((((((((......)....((.....)))))))))...))))))))((((((....))))))... ( -41.70) >consensus CGUGGAGCAUGCACUGAUUGACUUCAAGUACAUGGAAAAGAUAUUCCUAAAUGUAACGCUGCGCGAGAACAAAUGCAACAAGCGCAGCGUCGAAUUCCGCAUGCUUGGCAACGAGCAGUU .((.((((((((.....(((((......(((((((((......))))...)))))..((((((...................))))))))))).....)))))))).))........... (-20.38 = -21.66 + 1.28)

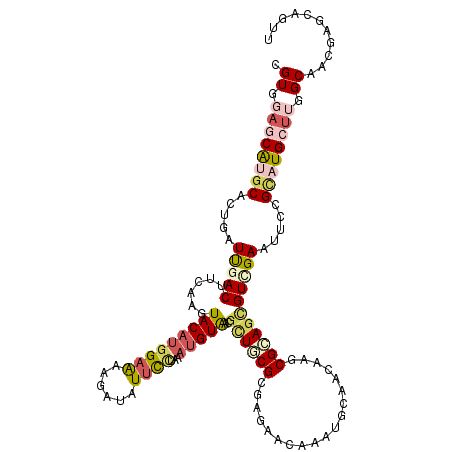
| Location | 7,657,013 – 7,657,133 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -21.05 |
| Energy contribution | -21.24 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7657013 120 + 20766785 AUAUUCCUGAAUGUAACGCUUCGAGAGGACAAGUGCAACAAGCGGAGCGUGGAGUUCCGUAUGCUGGGCAACGAGCAGUUUUCGCUCAAGAACAGGAAUUACUUCCAGGUAAUCAUUGCC ..(((((((........(.(((....)))).((((.(((..(((((((.....))))))).((((.(....).)))))))..))))......)))))))........((((.....)))) ( -34.80) >DroVir_CAF1 157 118 + 1 AUAUUCCUAAAUGUAACGCUACGCGAGAACAAAUGCAACAAGCGCAGCGUCGAAUUCCGCAUGCUGGGCAACGAGCAGUUUUCCCUGAAGAACAGGAAAUACUUUCAGGCAAGUGG--GC .................(((.(((((((((...(((.....((.((((((((.....)).)))))).)).....)))))))))((((((....((......))))))))...))))--)) ( -30.90) >DroGri_CAF1 157 118 + 1 AUAUUCUUAACUGUGUCGCUGCGCGAGAAUAAAUGCAAUAAGCGCAGCGUCGAAUUUCGCAUGCUUGGCAACGAGCAAUUCUCGCUAAAAAACAGGAAAUAUUUUCAGGCAAGUAA--CC ..............((..((((((((((((...(((.......)))(((........))).((((((....)))))))))))))).........((((....))))..)).))..)--). ( -32.90) >DroMoj_CAF1 157 115 + 1 AUAUUCCUAAAUGUAACGUUGCGCGAAAGUAAAUGCAACAAGCGCAGUGUCGAAUUUCGCAUGCUUGGCAACGAGCAGUUCUCCCUAAAGAAUAGAAAAUACUUUCAGGCAAGUA----- ...............((.((((..(((((((..........(((.(((.....))).))).((((((....))))))(((((......)))))......)))))))..)))))).----- ( -28.10) >DroAna_CAF1 157 117 + 1 AUCUUCCUGAACGUAACGCUGCGGGAGGACAAGUGCAAUAAGCGCAGUGUGGAGUUCAGGAUGCUUGGCAACGAGCAAUUCUCUCUGAAGAACCGAAAAUAUUUUCAGGUAAGAGGA--- ....((((((((...((((((((......)....((.....)))))))))...))))))))((((((....)))))).(((((((((((((..........)))))))...))))))--- ( -45.20) >DroPer_CAF1 157 117 + 1 AUCUUCCUGAACGUGACCCUGCGAGAGGACAAGUGCAACAAGAGGAGUGUCGAAUUCCGCAUGCUUGGAAACGAGCAGUUCUCCCUAAAGAACAGGAAAUACUUUCAGGUGAGUCAA--- .((..((((((.(((..((((...((((((...........(.(((((.....))))).).((((((....)))))))))))).(....)..))))...))).)))))).)).....--- ( -37.00) >consensus AUAUUCCUAAAUGUAACGCUGCGCGAGAACAAAUGCAACAAGCGCAGCGUCGAAUUCCGCAUGCUUGGCAACGAGCAGUUCUCCCUAAAGAACAGGAAAUACUUUCAGGCAAGUAA____ .....((((((.(((((((((((...................))))))))....((((...((((((....))))))(((((......))))).)))).)))))).)))........... (-21.05 = -21.24 + 0.20)

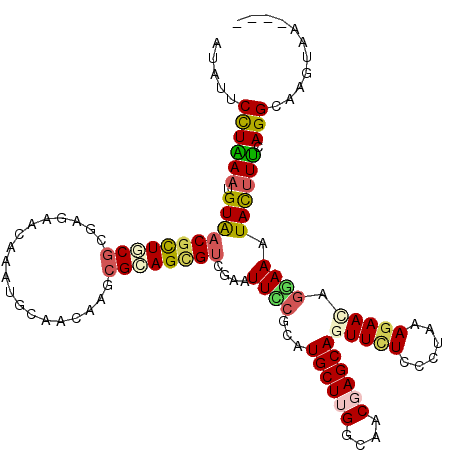
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:15 2006