
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,651,437 – 7,651,542 |
| Length | 105 |
| Max. P | 0.991578 |

| Location | 7,651,437 – 7,651,542 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.49 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -17.85 |
| Energy contribution | -20.27 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
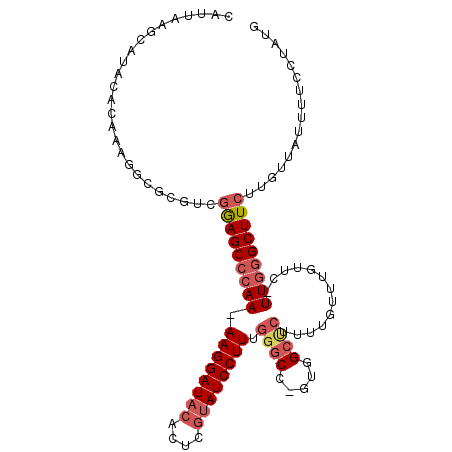
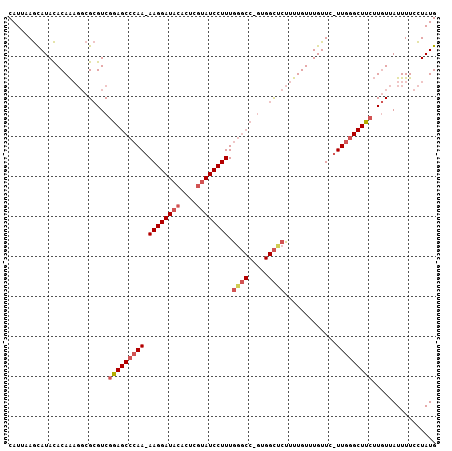
>2R_DroMel_CAF1 7651437 105 + 20766785 CAUUAAGCAUACACAAAGGCGCGUCGGAGCCCAA-AAGGAUACACUCGUAUCCUUUGGGCCAGUGGCUCUUUUGUUUGUUC-UUGGGCUUCUUGUUAUUUUCCUAUG ..(((((.(((.(((((((.((....(.((((((-(.((((((....))))))))))))))....)).))))))).))).)-))))..................... ( -33.60) >DroSim_CAF1 74019 105 + 1 CAUUAAGUAUACACAAAGGCGCGUCGGAGCCCAA-AAGGAUACACUCGUAUCCUUUGGGCCUGUGGCCCUUUUGUUUGUUC-UUGGGCUUCUUGUUAUUUUCCUAUG ..(((((.(((.(((((((.((..(((.((((((-(.((((((....))))))))))))))))..)).))))))).))).)-))))..................... ( -36.70) >DroAna_CAF1 97843 92 + 1 CAUUAAACAUAUACAG----GCGUCGAAGCCCAAAAAGGAUAUGCCC--AUCCUUUU---------UUCGGUUGUUUGUUCCUUUUGCUUAGUUUAAUUUUCUUAUG .(((((((.......(----((......)))..(((((((.(((..(--(.((....---------...)).))..)))))))))).....)))))))......... ( -17.50) >consensus CAUUAAGCAUACACAAAGGCGCGUCGGAGCCCAA_AAGGAUACACUCGUAUCCUUUGGGCC_GUGGCUCUUUUGUUUGUUC_UUGGGCUUCUUGUUAUUUUCCUAUG .........................(((((((((.((((((((....)))))))).((((.....)))).............)))))))))................ (-17.85 = -20.27 + 2.42)

| Location | 7,651,437 – 7,651,542 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.49 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -14.02 |
| Energy contribution | -15.83 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
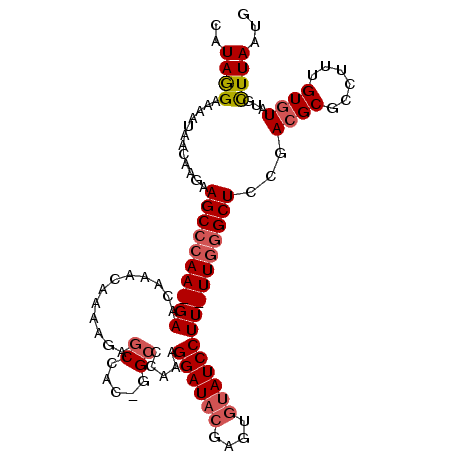
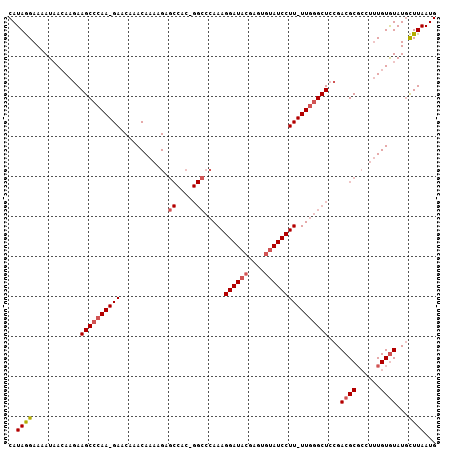
>2R_DroMel_CAF1 7651437 105 - 20766785 CAUAGGAAAAUAACAAGAAGCCCAA-GAACAAACAAAAGAGCCACUGGCCCAAAGGAUACGAGUGUAUCCUU-UUGGGCUCCGACGCGCCUUUGUGUAUGCUUAAUG .......................((-(.....(((((.(.((....((((((((((((((....)))))).)-))))))).....)).).))))).....))).... ( -27.50) >DroSim_CAF1 74019 105 - 1 CAUAGGAAAAUAACAAGAAGCCCAA-GAACAAACAAAAGGGCCACAGGCCCAAAGGAUACGAGUGUAUCCUU-UUGGGCUCCGACGCGCCUUUGUGUAUACUUAAUG ..((((..........(.(((((((-((..........(((((...)))))...((((((....))))))))-))))))).).(((((....)))))...))))... ( -34.60) >DroAna_CAF1 97843 92 - 1 CAUAAGAAAAUUAAACUAAGCAAAAGGAACAAACAACCGAA---------AAAAGGAU--GGGCAUAUCCUUUUUGGGCUUCGACGC----CUGUAUAUGUUUAAUG .........(((((((((.((...................(---------((((((((--(....))))))))))((((......))----)))).)).))))))). ( -19.80) >consensus CAUAGGAAAAUAACAAGAAGCCCAA_GAACAAACAAAAGAGCCAC_GGCCCAAAGGAUACGAGUGUAUCCUU_UUGGGCUCCGACGCGCCUUUGUGUAUGCUUAAUG ..((((............((((((((((............((.....)).....((((((....))))))))))))))))...((((......))))...))))... (-14.02 = -15.83 + 1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:13 2006