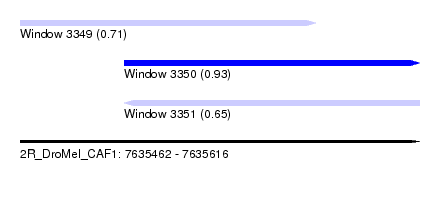
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,635,462 – 7,635,616 |
| Length | 154 |
| Max. P | 0.929154 |

| Location | 7,635,462 – 7,635,576 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -16.09 |
| Energy contribution | -16.78 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7635462 114 + 20766785 CCUUCGUUCCGGCGGGGCAUUUCCCCAGUUCGACUCGAAUU-----UGGGUAAUCCCAUUUAGCUGUC-AUCAGCUGUGGCUCCACUAAUCCCAACAACAGCCGAGGAUUCAAUCACUGA .....((((((((.(((.(((.(((.((((((...))))))-----.))).))))))....(((((..-..)))))(((....)))..............)))).))))........... ( -33.70) >DroSec_CAF1 80325 114 + 1 CCUUCGUUCCGGUGGGGCAUUUCCCCAGUUCGACUCGAAUU-----UGGGUAAUCCCAUUUAGCUGUC-AUCAGCUGUGGCUCCUCUAAUCCCAACUACAGCCGAGGAUUCAAUCACUGA .........((((((((.(((.(((.((((((...))))))-----.))).))))))...((((((..-..))))))((..(((((.................)))))..))..))))). ( -34.83) >DroYak_CAF1 83373 114 + 1 CCUUCGUGCUGGCGGGGCAUUUCCCCAGUUCGACUCGAAUU-----UGGGUAAUCCCAUUUAGCUGUC-AUCAGCUGUGGCUGCCCUAAUCCCAAGUACACCCGAGGAUUCAAUCACUGA ((((.(((((((.((((((...(((.((((((...))))))-----.))).....((((..(((((..-..))))))))).))))))....)).)))))....))))............. ( -36.50) >DroAna_CAF1 79980 97 + 1 CCAUCGCU-UGGCGGGGCAUUUCCCAGCUCCAAGCCGAAUAAGGACUUGGUAAUCCCUAUUACCUGUCUAUCUAGUCAGUUUCC---------------------UGAUUGCAUCA-UGA .....(((-(((((((((........)))))..)))))....((((..((((((....)))))).))))....((((((....)---------------------)))))))....-... ( -28.40) >consensus CCUUCGUUCCGGCGGGGCAUUUCCCCAGUUCGACUCGAAUU_____UGGGUAAUCCCAUUUAGCUGUC_AUCAGCUGUGGCUCC_CUAAUCCCAACUACAGCCGAGGAUUCAAUCACUGA .(((((........(((.(((..((((((((.....))))......)))).))))))...((((((.....)))))).........................)))))............. (-16.09 = -16.78 + 0.69)

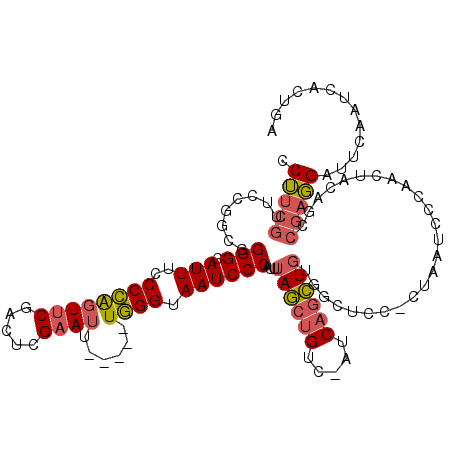
| Location | 7,635,502 – 7,635,616 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -32.06 |
| Energy contribution | -31.73 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7635502 114 + 20766785 UUGGGUAAUCCCAUUUAGCUGUCAUCAGCUGUGGCUCCACUAAUCCCAACAACAGCCGAGGAUUCAAUCACUGAAUGCACUUUUCACUGGGCUUACCCGGAAUCGCGUUGUGUU (((((...........(((((....)))))(((....)))....)))))..(((((((((.(((((.....))))).)........(((((....)))))..))).)))))... ( -34.20) >DroSec_CAF1 80365 114 + 1 UUGGGUAAUCCCAUUUAGCUGUCAUCAGCUGUGGCUCCUCUAAUCCCAACUACAGCCGAGGAUUCAAUCACUGAAUGCACUUUUCACUGGGCUUACCCAGAAUCGCGUUGUGCU (((((.....((((..(((((....)))))))))..........))))).((((((((((.(((((.....))))).)........(((((....)))))..))).)))))).. ( -34.46) >DroYak_CAF1 83413 114 + 1 UUGGGUAAUCCCAUUUAGCUGUCAUCAGCUGUGGCUGCCCUAAUCCCAAGUACACCCGAGGAUUCAAUCACUGAAUGCACUUCUCGCUGGGCUUACCCGGAAUCGUUGUGUUAU ..(((((...((((..(((((....))))))))).)))))...........((((.((((.(((((.....))))).)........(((((....)))))..)))..))))... ( -32.60) >consensus UUGGGUAAUCCCAUUUAGCUGUCAUCAGCUGUGGCUCCACUAAUCCCAACUACAGCCGAGGAUUCAAUCACUGAAUGCACUUUUCACUGGGCUUACCCGGAAUCGCGUUGUGAU (((((.....((((..(((((....)))))))))..........)))))..(((((((((.(((((.....))))).)........(((((....)))))..))).)))))... (-32.06 = -31.73 + -0.33)

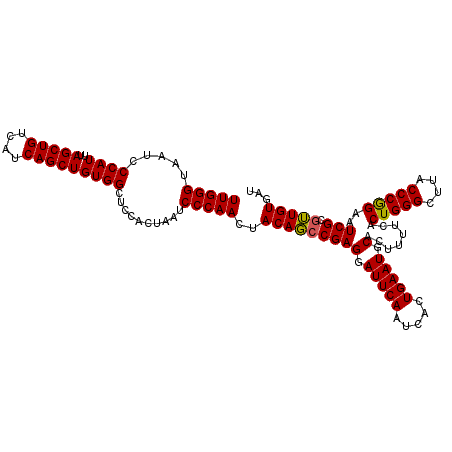

| Location | 7,635,502 – 7,635,616 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -32.38 |
| Energy contribution | -31.93 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7635502 114 - 20766785 AACACAACGCGAUUCCGGGUAAGCCCAGUGAAAAGUGCAUUCAGUGAUUGAAUCCUCGGCUGUUGUUGGGAUUAGUGGAGCCACAGCUGAUGACAGCUAAAUGGGAUUACCCAA ................((((((.((((......(((.(((.((((.....(((((.((((....))))))))).(((....))).)))))))...)))...)))).)))))).. ( -36.30) >DroSec_CAF1 80365 114 - 1 AGCACAACGCGAUUCUGGGUAAGCCCAGUGAAAAGUGCAUUCAGUGAUUGAAUCCUCGGCUGUAGUUGGGAUUAGAGGAGCCACAGCUGAUGACAGCUAAAUGGGAUUACCCAA .((.....)).....(((((((.((((.......(((..(((.....((.(((((.((((....))))))))).)).))).)))(((((....)))))...)))).))))))). ( -35.00) >DroYak_CAF1 83413 114 - 1 AUAACACAACGAUUCCGGGUAAGCCCAGCGAGAAGUGCAUUCAGUGAUUGAAUCCUCGGGUGUACUUGGGAUUAGGGCAGCCACAGCUGAUGACAGCUAAAUGGGAUUACCCAA ................((((((((((......((((((((((((.(((...))))).)))))))))).......))))..(((.(((((....)))))...)))..)))))).. ( -36.32) >consensus AACACAACGCGAUUCCGGGUAAGCCCAGUGAAAAGUGCAUUCAGUGAUUGAAUCCUCGGCUGUAGUUGGGAUUAGAGGAGCCACAGCUGAUGACAGCUAAAUGGGAUUACCCAA ................((((((.((((.......(((......((.....(((((.((((....)))))))))......)))))(((((....)))))...)))).)))))).. (-32.38 = -31.93 + -0.44)

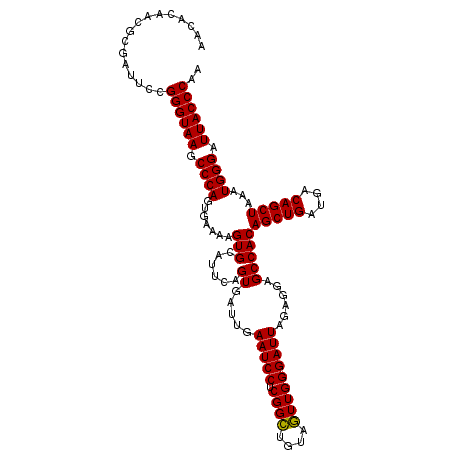
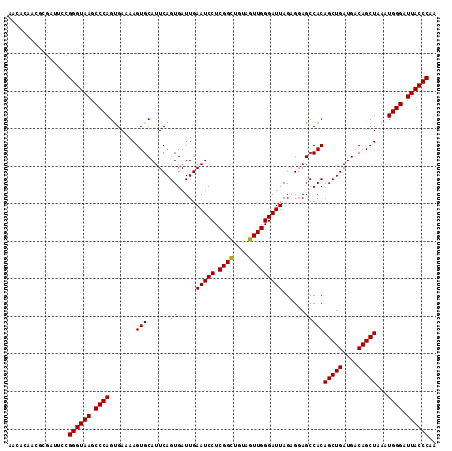
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:11 2006