| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,594,032 – 7,594,134 |
| Length | 102 |
| Max. P | 0.988322 |

| Location | 7,594,032 – 7,594,134 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -13.22 |
| Energy contribution | -13.47 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
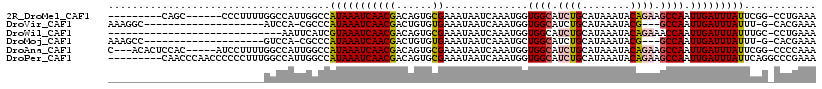
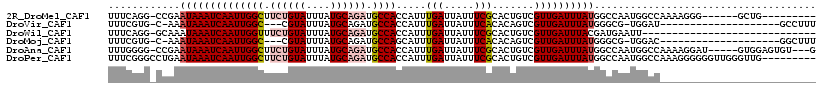
>2R_DroMel_CAF1 7594032 102 + 20766785 ---------CAGC------CCCUUUUGGCCAUUGGCCAUAAAUCAACGACAGUGCGAAAUAAUCAAAUGGUGGCAUCUGCAUAAAUACAGAAGCCAAUUGAUUUAUUCGG-CCUGAAA ---------....------......(((((...)))))...........(((..((((..((((((....((((.((((........)))).)))).))))))..)))).-.)))... ( -28.40) >DroVir_CAF1 55697 92 + 1 AAAGGC--------------------AUCCA-CGCCCAUAAAUCAACGACUGUGUGAAAUAAUCAAAUGGUGGCAUCUGCAUAAAUACG---GCCAAUUGAUUUAUUU-G-CACGAAA ...(((--------------------.....-.)))..............((((..((..((((((.((((.(.((........)).).---)))).))))))..)).-.-))))... ( -21.00) >DroWil_CAF1 53330 88 + 1 -----------------------------AAUUCAUCGUAAAUCAACGACAGUGCGAAAUAAUCAAAUGGUGGCAUCUGCAUAAAUACAGAAACCAAUUGAUUUAUUUGC-CCUGAAA -----------------------------......((((......))))(((.(((((..((((((.((((....((((........)))).)))).))))))..)))))-.)))... ( -23.60) >DroMoj_CAF1 67600 92 + 1 AAAGCC--------------------GUCCA-CGCCCAUAAAUCAACGACUGUGUGAAAUAAUCAAAUGCUGGCAUCUGCAUAAAUACG---GCCAAUUGAUUUAUUU-G-CACGAAA ......--------------------.....-.((..(((((((((...((((((((.....))..((((........))))..)))))---)....)))))))))..-)-)...... ( -16.90) >DroAna_CAF1 37698 109 + 1 C---ACACUCCAC-----AUCCUUUUGGCCAUUGGCCAUAAAUCAACGACAGUGCGAAAUAAUCAAAUGGUGGCAUCUGCAUAAAUACAGAAGCCAAUUGAUUUAUUCGG-CCCCAAA .---.((((....-----.......(((((...)))))............))))((((..((((((....((((.((((........)))).)))).))))))..)))).-....... ( -27.00) >DroPer_CAF1 36198 109 + 1 ---------CAACCCAACCCCCCUUUGGCCAUUGGCCAUAAAUCAACGACAGUGCGAAAUAAUCAAAUGGUGGCAUCUGCAUAAAUACAGAAGCCAAUUGAUUUAUUCAGGCCCGAAA ---------....((((.......)))).....(((((((((((((((......))..............((((.((((........)))).)))).)))))))))...))))..... ( -25.00) >consensus __________________________GGCCAUUGGCCAUAAAUCAACGACAGUGCGAAAUAAUCAAAUGGUGGCAUCUGCAUAAAUACAGAAGCCAAUUGAUUUAUUCGG_CCCGAAA .....................................(((((((((((......))..............((((.((((........)))).)))).)))))))))............ (-13.22 = -13.47 + 0.25)
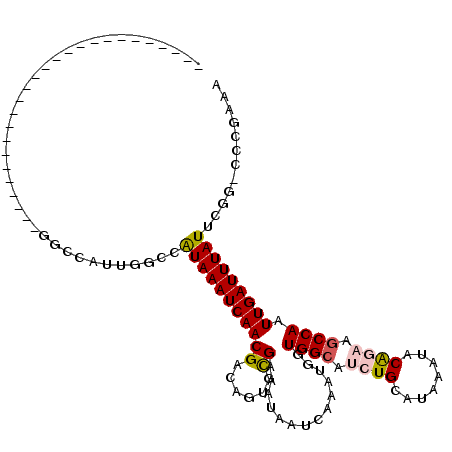
| Location | 7,594,032 – 7,594,134 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -16.78 |
| Energy contribution | -17.58 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
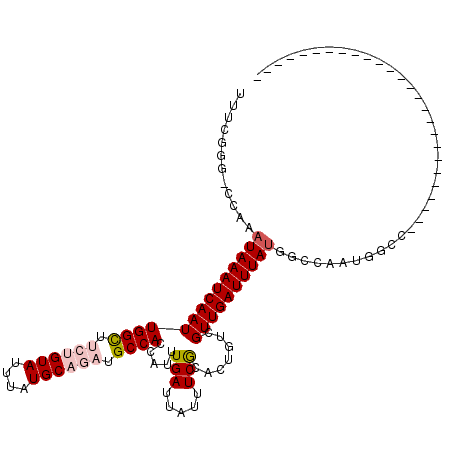
>2R_DroMel_CAF1 7594032 102 - 20766785 UUUCAGG-CCGAAUAAAUCAAUUGGCUUCUGUAUUUAUGCAGAUGCCACCAUUUGAUUAUUUCGCACUGUCGUUGAUUUAUGGCCAAUGGCCAAAAGGG------GCUG--------- .....((-((..((((((((((((((.((((((....)))))).))))......(((...........)))))))))))))))))...((((......)------))).--------- ( -33.60) >DroVir_CAF1 55697 92 - 1 UUUCGUG-C-AAAUAAAUCAAUUGGC---CGUAUUUAUGCAGAUGCCACCAUUUGAUUAUUUCACACAGUCGUUGAUUUAUGGGCG-UGGAU--------------------GCCUUU .((((((-(-..((((((((((.(((---.((.......((((((....))))))..........)).)))))))))))))..)))-)))).--------------------...... ( -21.63) >DroWil_CAF1 53330 88 - 1 UUUCAGG-GCAAAUAAAUCAAUUGGUUUCUGUAUUUAUGCAGAUGCCACCAUUUGAUUAUUUCGCACUGUCGUUGAUUUACGAUGAAUU----------------------------- ...(((.-((((((((.((((.((((.((((((....))))))....)))).)))))))))).)).)))((((((.....))))))...----------------------------- ( -25.10) >DroMoj_CAF1 67600 92 - 1 UUUCGUG-C-AAAUAAAUCAAUUGGC---CGUAUUUAUGCAGAUGCCAGCAUUUGAUUAUUUCACACAGUCGUUGAUUUAUGGGCG-UGGAC--------------------GGCUUU .((((((-(-..((((((((((.(((---.((.......((((((....))))))..........)).)))))))))))))..)))-)))).--------------------...... ( -23.23) >DroAna_CAF1 37698 109 - 1 UUUGGGG-CCGAAUAAAUCAAUUGGCUUCUGUAUUUAUGCAGAUGCCACCAUUUGAUUAUUUCGCACUGUCGUUGAUUUAUGGCCAAUGGCCAAAAGGAU-----GUGGAGUGU---G .......-..(((..((((((.((((.((((((....)))))).))))....))))))..)))(((((.(((((..(((.(((((...)))))))).)))-----).).)))))---. ( -33.10) >DroPer_CAF1 36198 109 - 1 UUUCGGGCCUGAAUAAAUCAAUUGGCUUCUGUAUUUAUGCAGAUGCCACCAUUUGAUUAUUUCGCACUGUCGUUGAUUUAUGGCCAAUGGCCAAAGGGGGGUUGGGUUG--------- ..((.(((((..((((((((((((((.((((((....)))))).))))......(((...........)))))))))))))((((...))))......))))).))...--------- ( -33.50) >consensus UUUCGGG_CCAAAUAAAUCAAUUGGCUUCUGUAUUUAUGCAGAUGCCACCAUUUGAUUAUUUCGCACUGUCGUUGAUUUAUGGCCAAUGGCC__________________________ ............((((((((((((((.((((((....)))))).)))).....(((.....))).......))))))))))..................................... (-16.78 = -17.58 + 0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:00 2006