
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,585,286 – 7,585,413 |
| Length | 127 |
| Max. P | 0.994450 |

| Location | 7,585,286 – 7,585,390 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -16.00 |
| Consensus MFE | -10.55 |
| Energy contribution | -10.18 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
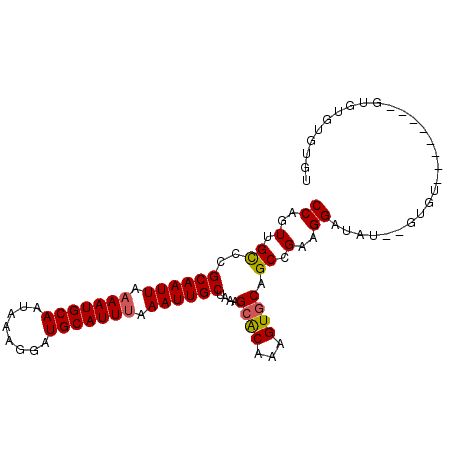
>2R_DroMel_CAF1 7585286 104 + 20766785 CGUGCCAGGAUCCCUAUUCCUUCCCCACC---G----CACACACACACACACGCACACAU--AUAUCCUUCGGCUGCACUUUGUGCUUUAGCAAUUUAAAUGCAUCCUUUAUU .((((.((((.......))))........---(----......)........))))....--.........((.((((.((..(((....)))....)).)))).))...... ( -16.30) >DroSec_CAF1 31190 96 + 1 CGUGCCAGGAUCCCUAUUCCUUCCCCACC---G----CACUCACACAC--------ACAC--AUAUCCUUCGGCUGCACUUUGUGCUUUAGCAAUUUUAAUGCAUCCUUUAUU .((((.((((.......))))........---)----)))........--------....--.........((.((((.....(((....))).......)))).))...... ( -15.00) >DroSim_CAF1 31936 88 + 1 CGUGCCAGGAUCCCUAUUCCUUCCC---------------ACACACAC--------ACAC--AUAUCCUUCGGCUGCACUUUGUGCUUUAGCAAUUUAAAUGCAUCCUUUAUU .(((..((((.......))))...)---------------))......--------....--.........((.((((.((..(((....)))....)).)))).))...... ( -15.80) >DroYak_CAF1 31756 113 + 1 CGUGCCAGGAUCCCUCUUCCUUUCCCUUUCCGUUCUCCUUGCACACACAGACACACAAGCAGAUAUCCUUCGGCUGUACUUUGCGCUUUAGCAAUUUAAAUGCAUCCUUUAUU .((((.((((.........................)))).))))..........(((.((.((......)).)))))...((((......))))................... ( -16.91) >consensus CGUGCCAGGAUCCCUAUUCCUUCCCCACC___G____CACACACACAC________ACAC__AUAUCCUUCGGCUGCACUUUGUGCUUUAGCAAUUUAAAUGCAUCCUUUAUU ......((((.......))))..................................................((.((((..((((......))))......)))).))...... (-10.55 = -10.18 + -0.38)

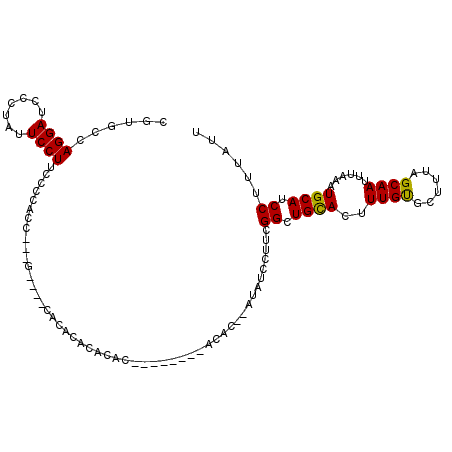
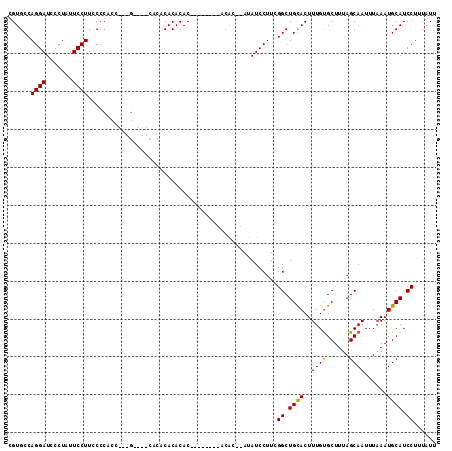
| Location | 7,585,317 – 7,585,413 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -18.83 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7585317 96 + 20766785 ACACACACACACACGCACACAU--AUAUCCUUCGGCUGCACUUUGUGCUUUAGCAAUUUAAAUGCAUCCUUUAUUGCAUUUUAAUUGCGGGCAACUGG ..............(((((...--.....(....)........)))))....((((((.(((((((........))))))).))))))((....)).. ( -23.19) >DroSec_CAF1 31221 88 + 1 ACUCACACAC--------ACAC--AUAUCCUUCGGCUGCACUUUGUGCUUUAGCAAUUUUAAUGCAUCCUUUAUUGCAUUUUAAUUGCGAACAACUGG ..........--------....--........(((..((((...))))....((((((..((((((........))))))..))))))......))). ( -18.70) >DroSim_CAF1 31961 86 + 1 --ACACACAC--------ACAC--AUAUCCUUCGGCUGCACUUUGUGCUUUAGCAAUUUAAAUGCAUCCUUUAUUGCAUUUUAAUUGCGGGCAACUGG --........--------....--....((....(((((((...))))....((((((.(((((((........))))))).)))))).)))....)) ( -22.10) >DroYak_CAF1 31794 98 + 1 UUGCACACACAGACACACAAGCAGAUAUCCUUCGGCUGUACUUUGCGCUUUAGCAAUUUAAAUGCAUCCUUUAUUGCAUUUUAAUUGCAGGCAACUGG .........(((........((((((((.(....)..))).)))))((((..((((((.(((((((........))))))).))))))))))..))). ( -25.90) >consensus ACACACACAC________ACAC__AUAUCCUUCGGCUGCACUUUGUGCUUUAGCAAUUUAAAUGCAUCCUUUAUUGCAUUUUAAUUGCGGGCAACUGG ............................((....(((((((...))))....((((((.(((((((........))))))).)))))).)))....)) (-18.83 = -19.20 + 0.37)

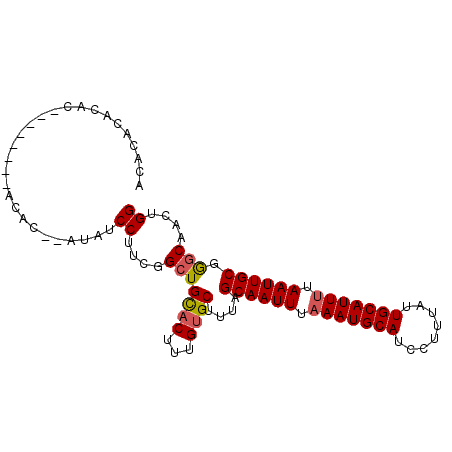
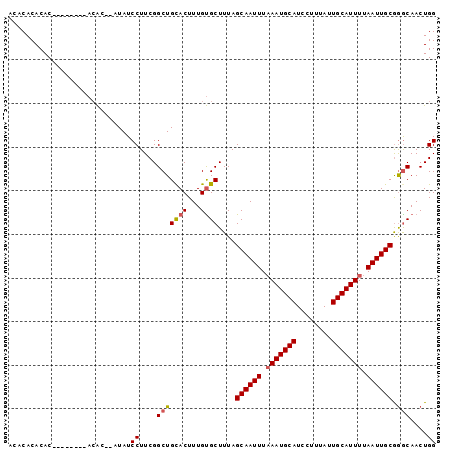
| Location | 7,585,317 – 7,585,413 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7585317 96 - 20766785 CCAGUUGCCCGCAAUUAAAAUGCAAUAAAGGAUGCAUUUAAAUUGCUAAAGCACAAAGUGCAGCCGAAGGAUAU--AUGUGUGCGUGUGUGUGUGUGU ((..(.((..((((((.(((((((........))))))).))))))....((((...)))).)).)..))((((--(((....)))))))........ ( -24.40) >DroSec_CAF1 31221 88 - 1 CCAGUUGUUCGCAAUUAAAAUGCAAUAAAGGAUGCAUUAAAAUUGCUAAAGCACAAAGUGCAGCCGAAGGAUAU--GUGU--------GUGUGUGAGU .......(((((((((..((((((........))))))..))))))....(((((..(..((..(....)...)--)..)--------.)))))))). ( -20.20) >DroSim_CAF1 31961 86 - 1 CCAGUUGCCCGCAAUUAAAAUGCAAUAAAGGAUGCAUUUAAAUUGCUAAAGCACAAAGUGCAGCCGAAGGAUAU--GUGU--------GUGUGUGU-- ..........((((((.(((((((........))))))).))))))....(((((..(..((..(....)...)--)..)--------...)))))-- ( -22.80) >DroYak_CAF1 31794 98 - 1 CCAGUUGCCUGCAAUUAAAAUGCAAUAAAGGAUGCAUUUAAAUUGCUAAAGCGCAAAGUACAGCCGAAGGAUAUCUGCUUGUGUGUCUGUGUGUGCAA ....((((..((((((.(((((((........))))))).))))))....(((((.((.(((..(((.((....))..)))..))))).))))))))) ( -26.10) >consensus CCAGUUGCCCGCAAUUAAAAUGCAAUAAAGGAUGCAUUUAAAUUGCUAAAGCACAAAGUGCAGCCGAAGGAUAU__GUGU________GUGUGUGUGU ((..(.((..((((((.(((((((........))))))).))))))....((((...)))).)).)..))............................ (-17.38 = -17.50 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:53 2006