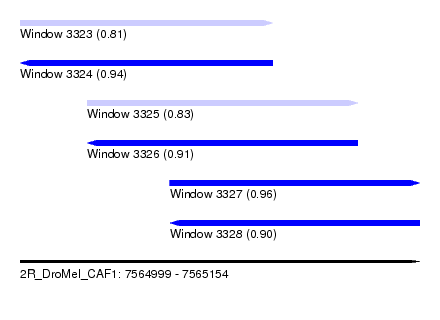
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,564,999 – 7,565,154 |
| Length | 155 |
| Max. P | 0.958517 |

| Location | 7,564,999 – 7,565,097 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -25.75 |
| Energy contribution | -26.53 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7564999 98 + 20766785 CGCUG--CCGAAGCGGC------UGCCAAUGAGGCCA-AUGCCAAUCCAA-AUAGAAAUCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAGGU .(.((--(((..(((((------.(((.....)))..-..)))..((...-...)).........))...))))).)((((((((((((.....))))))))..)))) ( -33.40) >DroVir_CAF1 12335 98 + 1 ---CA--CUGAAGCGGCUGUCGAGGUCAAUGAGGUCA-AUGCCAAUUCAA-AUGGCAAUGGCAA---CACUGGCGACAUCUGACUGCCCAUUCUGGGCAGCCUUAGAU ---..--((((..(((.((((...((((.((..((((-.(((((......-.))))).))))..---)).)))))))).))).((((((.....))))))..)))).. ( -36.50) >DroSec_CAF1 11421 102 + 1 CGCUUGCCCGAAGCGGC------GGCCAAUGAGGCCAAAUGCCCAUCCGAAAUAGAAUUCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAGGU ((((.((.....))(((------((((.....))))............(((......))).....)))...))))((....((((((((.....))))))))....)) ( -34.40) >DroSim_CAF1 11605 98 + 1 CGCUG--CCGAAGCGGC------GGCCAAUGAGGCCA-AUGCCAAUCCGA-AUAGAAAUCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAGGU .((((--(....)))))------((((.....)))).-..(((.......-.............((((...))))......((((((((.....))))))))...))) ( -37.60) >DroEre_CAF1 11364 98 + 1 CGCUG--CCGAAGCGGC------GGCCAAUGAGGCCA-AUGCCAAUCCAA-AUAGAAAUUCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUAAGGU .((((--(....)))))------((((.....)))).-..(((.......-.............((((...))))......((((((((.....))))))))...))) ( -37.60) >DroAna_CAF1 11076 98 + 1 CGCUG--CCGAGGUGGC------CGCCAACGAGGCCA-ACGCCAAUCCGA-GCGGCAAUGGCAACGCCAACGGCGACAUCUGACUGCCCAUACUGGGCAGCCUGAGGU (((((--(((.((((((------.(((.....)))..-..))))..))..-.))))..((((...))))..))))((.((.(.((((((.....)))))).).)).)) ( -40.00) >consensus CGCUG__CCGAAGCGGC______GGCCAAUGAGGCCA_AUGCCAAUCCAA_AUAGAAAUCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAGGU .(((.......)))(((......((((.....))))....)))...((................((((...))))......((((((((.....))))))))...)). (-25.75 = -26.53 + 0.78)

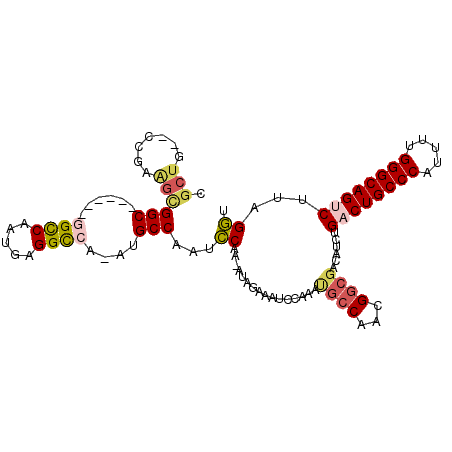
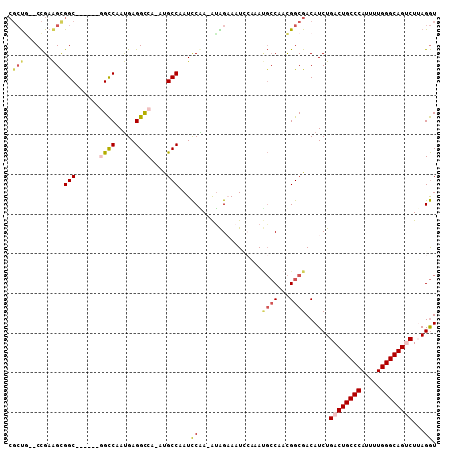
| Location | 7,564,999 – 7,565,097 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -31.72 |
| Energy contribution | -32.58 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7564999 98 - 20766785 ACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGAUUUCUAU-UUGGAUUGGCAU-UGGCCUCAUUGGCA------GCCGCUUCGG--CAGCG .((...((((((((.....)))))))).((((..((((.((.((((..(((....))-)..)).)).)).-))))..))))))..------((.((....)--).)). ( -42.10) >DroVir_CAF1 12335 98 - 1 AUCUAAGGCUGCCCAGAAUGGGCAGUCAGAUGUCGCCAGUG---UUGCCAUUGCCAU-UUGAAUUGGCAU-UGACCUCAUUGACCUCGACAGCCGCUUCAG--UG--- ......((((((((.....))))))))...(((((.(((((---..(.((.(((((.-......))))).-)).)..)))))....)))))..(((....)--))--- ( -35.60) >DroSec_CAF1 11421 102 - 1 ACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGAAUUCUAUUUCGGAUGGGCAUUUGGCCUCAUUGGCC------GCCGCUUCGGGCAAGCG ......((((((((.....))))))))((((((((((...)))(((((((((...))))))))).)))))))((((.....))))------(((......)))..... ( -43.00) >DroSim_CAF1 11605 98 - 1 ACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGAUUUCUAU-UCGGAUUGGCAU-UGGCCUCAUUGGCC------GCCGCUUCGG--CAGCG ......((((((((.....)))))))).(((((((((...)))((((((((....))-)))))).)))))-)((((.....))))------((.((....)--).)). ( -45.00) >DroEre_CAF1 11364 98 - 1 ACCUUAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGAAUUUCUAU-UUGGAUUGGCAU-UGGCCUCAUUGGCC------GCCGCUUCGG--CAGCG ......((((((((.....)))))))).(((((((((...)))(((..(((....))-)..))).)))))-)((((.....))))------((.((....)--).)). ( -44.10) >DroAna_CAF1 11076 98 - 1 ACCUCAGGCUGCCCAGUAUGGGCAGUCAGAUGUCGCCGUUGGCGUUGCCAUUGCCGC-UCGGAUUGGCGU-UGGCCUCGUUGGCG------GCCACCUCGG--CAGCG .((...((((((((.....)))))))).((.((.(((((..(((..((((.(((((.-......))))).-))))..)))..)))------)).)).))))--..... ( -51.80) >consensus ACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGAUUUCUAU_UCGGAUUGGCAU_UGGCCUCAUUGGCC______GCCGCUUCGG__CAGCG ......((((((((.....))))))))((((((((....))))))))............((((.((((....((((.....))))......)))).))))........ (-31.72 = -32.58 + 0.86)

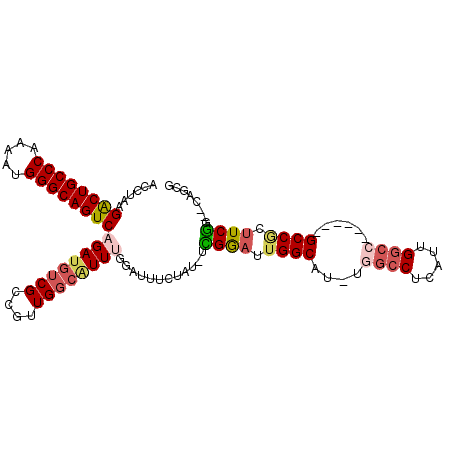
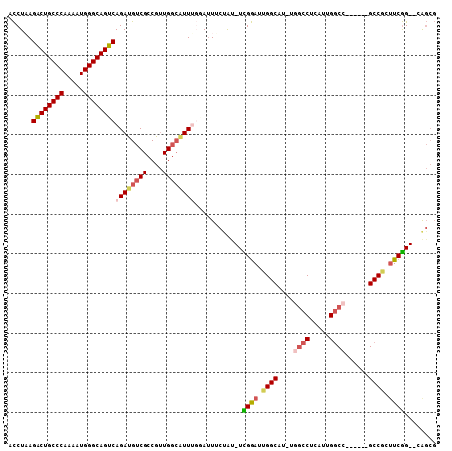
| Location | 7,565,025 – 7,565,130 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.87 |
| Mean single sequence MFE | -36.15 |
| Consensus MFE | -20.83 |
| Energy contribution | -21.22 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7565025 105 + 20766785 CCAAUGCCAAUCCAAAUAGAAAUCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAGGUAAAGUCUGCCGGCGGAAAACUAACGCAACUGGG (((.(((...((......))..........((..(((((((((((((((((((.....))))))))..))))...))).))))..)).........)))..))). ( -33.40) >DroVir_CAF1 12364 102 + 1 UCAAUGCCAAUUCAAAUGGCAAUGGCAA---CACUGGCGACAUCUGACUGCCCAUUCUGGGCAGCCUUAGAUGAAGCCUGGCCGCCAAUGAGUAGCCAAACUAAG ....(((((.......))))).((....---))..(((..(((((((((((((.....))))))..)))))))..)))((((..(......)..))))....... ( -39.50) >DroGri_CAF1 14104 102 + 1 UGAACGCCAAUGCGAAUGGCAAUGGCAA---CACUGGCGACAUCUGACUGCCCAUUUUAGGCAGCCUUAGAUGAAGCCUGGCCGCCAGGGAGUAGCCAAACUAAG .....((((.(((.....))).))))..---....(((..((((((((((((.......)))))..)))))))..)))((((..(......)..))))....... ( -38.10) >DroSim_CAF1 11631 105 + 1 CCAAUGCCAAUCCGAAUAGAAAUCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAGGUAAAGUCUGCCGGCGGAGGACUAACGCAACUGGG (((.(((...((......))..(((...((((...((((((((((((((((((.....))))))))..))))...))).)))))))..))).....)))..))). ( -35.50) >DroEre_CAF1 11390 105 + 1 CCAAUGCCAAUCCAAAUAGAAAUUCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUAAGGUGAAGUCUGCCGGCGGAGGACUAACGCAACUGGG (((.(((...(((.....(.....).....((..(((((((((((((((((((.....))))))))..))))...))).))))..)).))).....)))..))). ( -35.70) >DroYak_CAF1 11672 105 + 1 CCAAUGCCAAUCCAAAUAGAAACCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUCUGGGCAGUCUAAGGUGAAGUCUGCCGACGGAGGACUAAAGCAACUGGG ......................((((..(((.........(((((((((((((.....))))))))..))))).(((((.((...)).)))))...)))..)))) ( -34.70) >consensus CCAAUGCCAAUCCAAAUAGAAAUCCAAAUGCCAACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAGGUGAAGUCUGCCGGCGGAGGACUAACGCAACUGGG ...................................(((..(((((((((((((.....))))))))..)))))..)))........................... (-20.83 = -21.22 + 0.39)


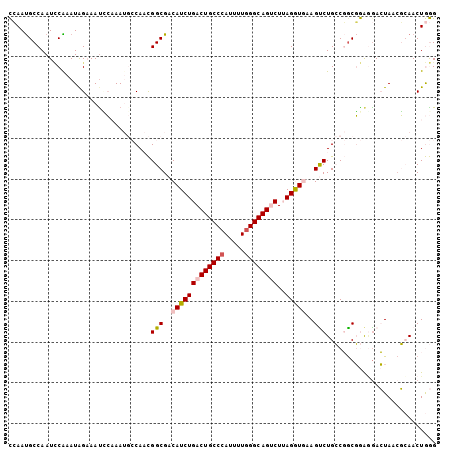
| Location | 7,565,025 – 7,565,130 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.87 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.40 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.42 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7565025 105 - 20766785 CCCAGUUGCGUUAGUUUUCCGCCGGCAGACUUUACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGAUUUCUAUUUGGAUUGGCAUUGG .(((((.((.........(((.((((.(((.....((..((((((((.....))))))))))..))))))).))).(((..(((....)))..)))..))))))) ( -36.50) >DroVir_CAF1 12364 102 - 1 CUUAGUUUGGCUACUCAUUGGCGGCCAGGCUUCAUCUAAGGCUGCCCAGAAUGGGCAGUCAGAUGUCGCCAGUG---UUGCCAUUGCCAUUUGAAUUGGCAUUGA .......((((....((((((((((........((((..((((((((.....))))))))))))))))))))))---..)))).(((((.......))))).... ( -44.70) >DroGri_CAF1 14104 102 - 1 CUUAGUUUGGCUACUCCCUGGCGGCCAGGCUUCAUCUAAGGCUGCCUAAAAUGGGCAGUCAGAUGUCGCCAGUG---UUGCCAUUGCCAUUCGCAUUGGCGUUCA .......((((..((....))..))))(((..(((((..((((((((.....)))))))))))))..)))....---..((((.(((.....))).))))..... ( -42.00) >DroSim_CAF1 11631 105 - 1 CCCAGUUGCGUUAGUCCUCCGCCGGCAGACUUUACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGAUUUCUAUUCGGAUUGGCAUUGG .(((((...((((((((.(((.((((.(((.....((..((((((((.....))))))))))..))))))).))).....((((....))))))))))))))))) ( -40.60) >DroEre_CAF1 11390 105 - 1 CCCAGUUGCGUUAGUCCUCCGCCGGCAGACUUCACCUUAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGAAUUUCUAUUUGGAUUGGCAUUGG .(((((...((((((((.(((.((((.(((.((......((((((((.....)))))))).)).))))))).))).....((((....))))))))))))))))) ( -39.20) >DroYak_CAF1 11672 105 - 1 CCCAGUUGCUUUAGUCCUCCGUCGGCAGACUUCACCUUAGACUGCCCAGAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGGUUUCUAUUUGGAUUGGCAUUGG .(((((.(((..(((((...(((....)))...((((..((((((((.....))))))))((((((((....))))))))))))........))))))))))))) ( -39.90) >consensus CCCAGUUGCGUUAGUCCUCCGCCGGCAGACUUCACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUUGGCAUUUGGAUUUCUAUUUGGAUUGGCAUUGG .(((((...((((((((...((((((.(((.((......((((((((.....)))))))).)).)))...))))))....((((....))))))))))))))))) (-25.98 = -26.40 + 0.42)
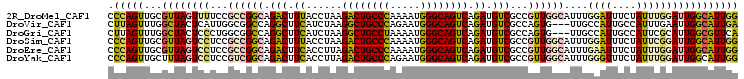
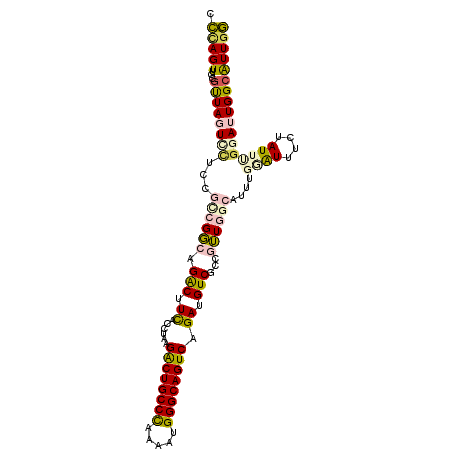
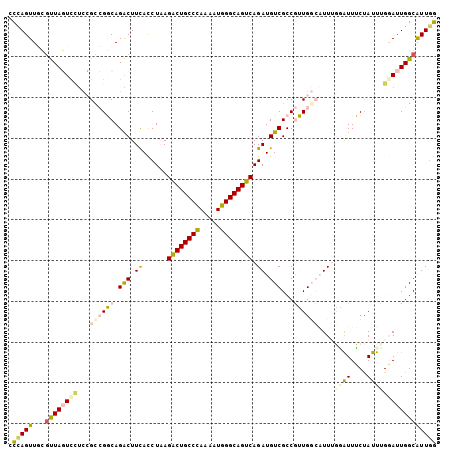
| Location | 7,565,057 – 7,565,154 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.79 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -20.83 |
| Energy contribution | -21.22 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7565057 97 + 20766785 AACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAGGUAAAGUCUGCCGGCGGAAAACUAACGCAACUGGGUGCGCCA----AUCAAAAUCAAGAGCCA ...((((.(((((((((((((.....))))))))...((((.....)))).(((.........)))....))))))))).----................. ( -36.50) >DroVir_CAF1 12393 99 + 1 ACUGGCGACAUCUGACUGCCCAUUCUGGGCAGCCUUAGAUGAAGCCUGGCCGCCAAUGAGUAGCCAAACUAAG-GCACAAUUUUAUCAA-ACGUUGUUUCA ...(((..(((((((((((((.....))))))..)))))))..)))((((..(......)..))))......(-(.(((((.((....)-).))))).)). ( -34.30) >DroGri_CAF1 14133 99 + 1 ACUGGCGACAUCUGACUGCCCAUUUUAGGCAGCCUUAGAUGAAGCCUGGCCGCCAGGGAGUAGCCAAACUAAG-GCACAAUUUUAUCAA-AUGAUGUUUCA .((((((.((((((((((((.......)))))..)))))))..((...))))))))(((((.(((.......)-)))).....((((..-..)))).))). ( -31.00) >DroSim_CAF1 11663 97 + 1 AACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAGGUAAAGUCUGCCGGCGGAGGACUAACGCAACUGGGUGCGCAA----AUCAAAAUCAAGAGCCA ...((((((((((((((((((.....))))))))..))))...))).)))(((.((.((....((((......))))...----.))....))....))). ( -35.30) >DroEre_CAF1 11422 97 + 1 AACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUAAGGUGAAGUCUGCCGGCGGAGGACUAACGCAACUGGGUGUGCAA----AUCAAAAUCGAGAGCCA ...(((.((((((((((((((.....))))))))....(((.(((((.((...)).)))))..)))....))))))....----.((......))..))). ( -34.70) >DroYak_CAF1 11704 97 + 1 AACGGCGACAUCUGACUGCCCAUUCUGGGCAGUCUAAGGUGAAGUCUGCCGACGGAGGACUAAAGCAACUGGGUGUGCAA----AUCAAAAUCAAGAGCCA ...(((..(((((((((((((.....))))))))..))))).(((((.((...)).)))))...(((.(.....))))..----.............))). ( -34.00) >consensus AACGGCGACAUCUGACUGCCCAUUUUGGGCAGUCUUAGGUGAAGUCUGCCGGCGGAGGACUAACGCAACUGGGUGCGCAA____AUCAAAAUCAAGAGCCA ...(((..(((((((((((((.....))))))))..)))))..)))....................................................... (-20.83 = -21.22 + 0.39)

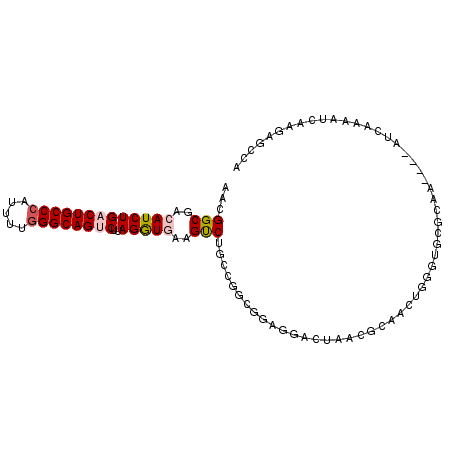
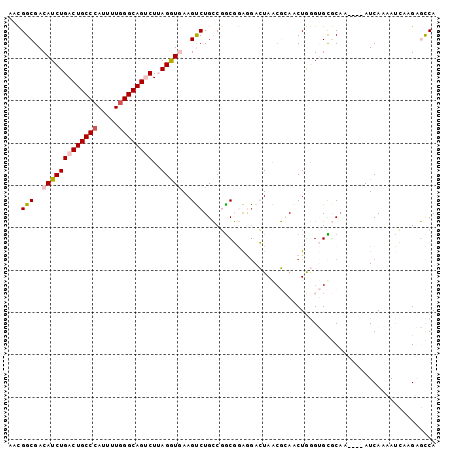
| Location | 7,565,057 – 7,565,154 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.79 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -20.22 |
| Energy contribution | -19.75 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7565057 97 - 20766785 UGGCUCUUGAUUUUGAU----UGGCGCACCCAGUUGCGUUAGUUUUCCGCCGGCAGACUUUACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUU .(((....((....(((----(((((((......)))))))))).)).)))(((.(((.....((..((((((((.....))))))))))..))))))... ( -36.90) >DroVir_CAF1 12393 99 - 1 UGAAACAACGU-UUGAUAAAAUUGUGC-CUUAGUUUGGCUACUCAUUGGCGGCCAGGCUUCAUCUAAGGCUGCCCAGAAUGGGCAGUCAGAUGUCGCCAGU .......((((-(((((........((-(((((((((((..((....))..))))))))......)))))(((((.....))))))))))))))....... ( -35.40) >DroGri_CAF1 14133 99 - 1 UGAAACAUCAU-UUGAUAAAAUUGUGC-CUUAGUUUGGCUACUCCCUGGCGGCCAGGCUUCAUCUAAGGCUGCCUAAAAUGGGCAGUCAGAUGUCGCCAGU .....(((.((-((....)))).))).-.......((((..((....))..))))(((..(((((..((((((((.....)))))))))))))..)))... ( -33.30) >DroSim_CAF1 11663 97 - 1 UGGCUCUUGAUUUUGAU----UUGCGCACCCAGUUGCGUUAGUCCUCCGCCGGCAGACUUUACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUU .(((....((....(((----(.(((((......))))).)))).)).)))(((.(((.....((..((((((((.....))))))))))..))))))... ( -34.10) >DroEre_CAF1 11422 97 - 1 UGGCUCUCGAUUUUGAU----UUGCACACCCAGUUGCGUUAGUCCUCCGCCGGCAGACUUCACCUUAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUU .(((....((((.....----.((((........))))..))))....)))(((.(((.((......((((((((.....)))))))).)).))))))... ( -31.20) >DroYak_CAF1 11704 97 - 1 UGGCUCUUGAUUUUGAU----UUGCACACCCAGUUGCUUUAGUCCUCCGUCGGCAGACUUCACCUUAGACUGCCCAGAAUGGGCAGUCAGAUGUCGCCGUU .(((....(((...(((----(.(((........)))...))))....)))(((((....)......((((((((.....))))))))...)))))))... ( -29.30) >consensus UGGCUCUUGAUUUUGAU____UUGCGCACCCAGUUGCGUUAGUCCUCCGCCGGCAGACUUCACCUAAGACUGCCCAAAAUGGGCAGUCAGAUGUCGCCGUU ...................................................(((.(((.((......((((((((.....)))))))).)).))))))... (-20.22 = -19.75 + -0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:49 2006