
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,559,298 – 7,559,391 |
| Length | 93 |
| Max. P | 0.944051 |

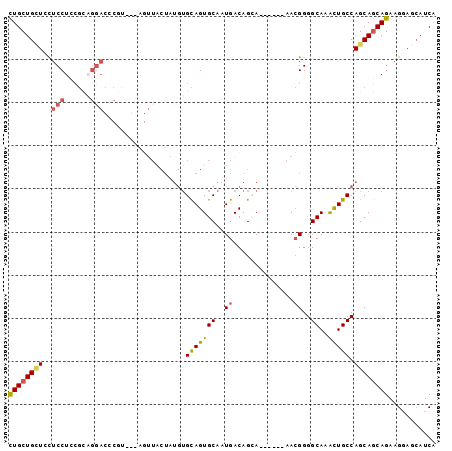
| Location | 7,559,298 – 7,559,391 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -17.12 |
| Energy contribution | -17.46 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
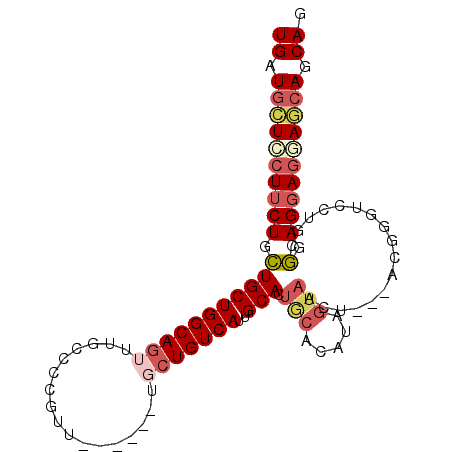
>2R_DroMel_CAF1 7559298 93 + 20766785 CUGCUGCUUCUCCUCCGCAGGACACGUAGAAGUUACUAUGUGCAGUGCAAUGACAGCA------AACGGGGCAAACUGCCAGCAGCAGAAGCAGCAUCA .((((((((((.((.((((.(.(((((((......))))))))..)))..........------.....(((.....))).).)).))))))))))... ( -34.30) >DroSec_CAF1 5777 93 + 1 CUGCUGCUCCUCCUCCGCAGGACCCGUAGAAGUUACUAUGUGUAGUGCAAUGACAGAA------AACGGGGCAAACUGCCAGCAGCAGAAGGAGCAUCA ((((((((........(((..((.(((((......))))).))..)))..........------.....(((.....)))))))))))........... ( -28.40) >DroSim_CAF1 5913 93 + 1 CUGCUGCUCCUCCUCCGCAGGACCCGUAGAAGUUACUAUGUGCAGUGCAAUGACAGCA------AACGGGGCAAACUGCCAGCAGCAGAAGGAGCAUCA ..(.(((((((.(((((.......(((((......)))))(((.((......)).)))------..)))))....((((.....)))).))))))).). ( -27.40) >DroEre_CAF1 5761 90 + 1 CUGCUGCUGCUCCUUCGUAGGACCCGC---AGUUACUAUGUGCAGUGCAAUGACAGCA------AACGGGGCAAACUGCCAGCAGCAGAAGGAGCAUCA (((((((((.((((....))))...((---((((....(((.(..(((.......)))------....).))))))))))))))))))........... ( -32.10) >DroYak_CAF1 5808 76 + 1 CUGCUGGUCCUCCUCC-----------------UACUAUGUGCAGUGCAAUGACAGCA------AACGGGGCACAUUGCCAGCAGCAGAAGGAGCAUCA .....(((.(((((.(-----------------(.((.((.(((((((..((......------..))..))))..)))))..)).)).))))).))). ( -21.90) >DroPer_CAF1 6001 81 + 1 UUGCUGUUGUUCCUGUGUUGGA------------------GGCAGUGCAAUGACAGGACUGCAACAAGGGGCAAGCUGCCAGCAACAGUAGCAAAAGCA .(((((((((((((.((((...------------------.(((((.(.......).))))))))))))(((.....)))))))))))))((....)). ( -29.00) >consensus CUGCUGCUCCUCCUCCGCAGGACCCGU___AGUUACUAUGUGCAGUGCAAUGACAGCA______AACGGGGCAAACUGCCAGCAGCAGAAGGAGCAUCA ((((((((..(((......)))...................(((((((..((..............))..))..))))).))))))))........... (-17.12 = -17.46 + 0.34)


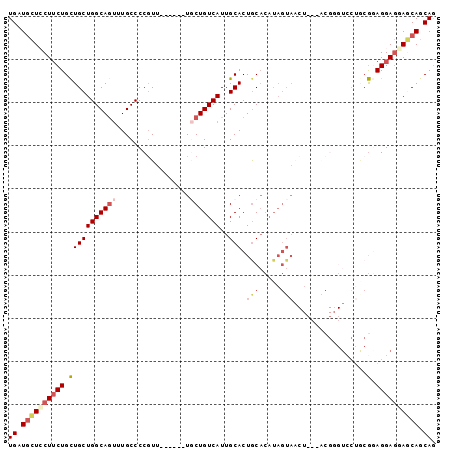
| Location | 7,559,298 – 7,559,391 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -17.45 |
| Energy contribution | -18.68 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7559298 93 - 20766785 UGAUGCUGCUUCUGCUGCUGGCAGUUUGCCCCGUU------UGCUGUCAUUGCACUGCACAUAGUAACUUCUACGUGUCCUGCGGAGGAGAAGCAGCAG ...((((((((((.((.((((((((..((...)).------.)))))))..(((..((((.(((......))).))))..)))).)).)))))))))). ( -41.10) >DroSec_CAF1 5777 93 - 1 UGAUGCUCCUUCUGCUGCUGGCAGUUUGCCCCGUU------UUCUGUCAUUGCACUACACAUAGUAACUUCUACGGGUCCUGCGGAGGAGGAGCAGCAG ((.((((((((((.((((.(((.....)))((((.------....((.((((.........)))).))....)))).....)))))))))))))).)). ( -31.90) >DroSim_CAF1 5913 93 - 1 UGAUGCUCCUUCUGCUGCUGGCAGUUUGCCCCGUU------UGCUGUCAUUGCACUGCACAUAGUAACUUCUACGGGUCCUGCGGAGGAGGAGCAGCAG ((.((((((((((.((((.(((.....)))(((((------((((((...(((...))).))))))).....)))).....)))))))))))))).)). ( -36.50) >DroEre_CAF1 5761 90 - 1 UGAUGCUCCUUCUGCUGCUGGCAGUUUGCCCCGUU------UGCUGUCAUUGCACUGCACAUAGUAACU---GCGGGUCCUACGAAGGAGCAGCAGCAG ((.((((((((((((.....)))((..(.((((((------((((((...(((...))).)))))))..---))))).)..))))))))))).)).... ( -33.80) >DroYak_CAF1 5808 76 - 1 UGAUGCUCCUUCUGCUGCUGGCAAUGUGCCCCGUU------UGCUGUCAUUGCACUGCACAUAGUA-----------------GGAGGAGGACCAGCAG ((.(.(((((((((((((..((((((.((......------.))...))))))...))....))))-----------------))))))).).)).... ( -28.10) >DroPer_CAF1 6001 81 - 1 UGCUUUUGCUACUGUUGCUGGCAGCUUGCCCCUUGUUGCAGUCCUGUCAUUGCACUGCC------------------UCCAACACAGGAACAACAGCAA ((((.(((...((((((..(((((............((((((......)))))))))))------------------..))..))))...))).)))). ( -22.30) >consensus UGAUGCUCCUUCUGCUGCUGGCAGUUUGCCCCGUU______UGCUGUCAUUGCACUGCACAUAGUAACU___ACGGGUCCUGCGGAGGAGGAGCAGCAG ((.((((((((((.(((((((((((.................)))))))..))).(((.....))).................).)))))))))).)). (-17.45 = -18.68 + 1.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:43 2006