
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,553,668 – 7,553,774 |
| Length | 106 |
| Max. P | 0.845639 |

| Location | 7,553,668 – 7,553,774 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.29 |
| Mean single sequence MFE | -42.33 |
| Consensus MFE | -26.73 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
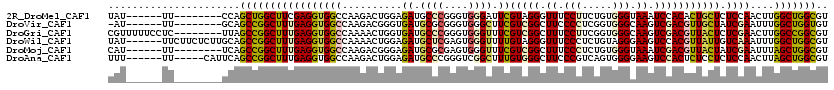
>2R_DroMel_CAF1 7553668 106 + 20766785 UAU------UU--------CCAGCUGGCUUCGAGGUGGCCAAGACUGGAGAUGCCCGGGUGGGAUUCGUAGGGUUUCCUUCUGUGGGUAAAUCCACACUGCUCUCCAACUUGGCUGGCGU .((------((--------((((((((((.......)))).)).))))))))((((((((.(((...((((((...))...((((((....))))))))))..))).)))))...))).. ( -37.60) >DroVir_CAF1 170 105 + 1 -AU------UU--------GCAGCCGGCUUUGAGGUGGCCAAGACGGGUGAUGCGCGGGUGGGCUUCGUCGGCUUCCCCUCGGUGGGCAAGUCGACGUUGCUAUCGAAUUUGGCUGGUGU -..------..--------.(((((((.((((((((((((....((.(.....).))....)))).(((((((((.(((.....))).)))))))))..))).))))).))))))).... ( -44.00) >DroGri_CAF1 173 112 + 1 CGUUUUUCCUC--------UUAGCCGGCUUUGAGGUGGCCAAAACUGGUGAUGCCCGGGUGGGUUUCGUCGGCUUUCCUUCGGUGGGCAAGUCGACGUUACUCUCGAACUUGGCCGGCGU ...........--------...(((((((((((((((((..........((.((((....)))).))((((((((.(((.....))).))))))))))))).)))))....))))))).. ( -47.10) >DroWil_CAF1 167 114 + 1 UAU------UUCUUCUCUUGCAGCCGGCUUUGAGGUGGCCAAAACUGGAGAUGCUCGAGUGGGUUUUGUAGGGUUUCCCUCUGUAGGGAAGUCCACGUUAUUGUCAAAUUUGGCUGGCGU ...------.............(((((((..((..((((....(((.(((...))).))).(((..(((.((..((((((....))))))..)))))..)))))))..)).))))))).. ( -38.20) >DroMoj_CAF1 170 106 + 1 CAU------UU--------UCAGCCGGCUUUGAGGUGGCCAAGACGGGAGAUGCGCGAGUGGGUUUCGUCGGCUUUCCCUCUGUGGGUAAAUCGACGUUACUAUCGAAUUUAGCUGGCGU ...------..--------...(((((((..((((.((((..((((..(..(((....)))..)..))))))))...))))..........((((........))))....))))))).. ( -38.40) >DroAna_CAF1 172 109 + 1 UUU------UU-----CAUUCAGCCGGCUUUGAGGUGGCCAAGACUGGAGAUGCCCGGGUCGGCUUUGUGGGCUUCCCGUCAGUGGGGAAGUCCACUCUCCUCUCCAACUUAGCUGGCGU ...------..-----......(((((((..(((.(((...(((..(((((.(((......)))...(((((((((((.......)))))))))))))))))))))).)))))))))).. ( -48.70) >consensus UAU______UU________UCAGCCGGCUUUGAGGUGGCCAAGACUGGAGAUGCCCGGGUGGGUUUCGUCGGCUUUCCCUCUGUGGGCAAGUCCACGUUACUCUCGAACUUGGCUGGCGU ......................(((((((((((((((((..........((.((((....)))).))(((((.((.(((.....))).)).))))))))))).))))....))))))).. (-26.73 = -26.90 + 0.17)
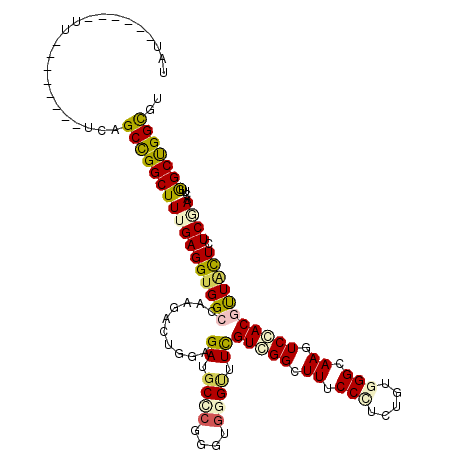
| Location | 7,553,668 – 7,553,774 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.29 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -18.41 |
| Energy contribution | -18.13 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
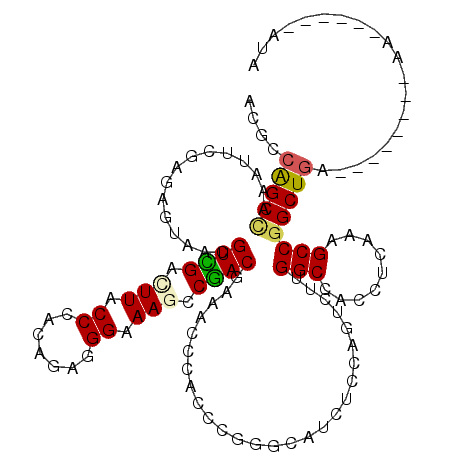
>2R_DroMel_CAF1 7553668 106 - 20766785 ACGCCAGCCAAGUUGGAGAGCAGUGUGGAUUUACCCACAGAAGGAAACCCUACGAAUCCCACCCGGGCAUCUCCAGUCUUGGCCACCUCGAAGCCAGCUGG--------AA------AUA ...(((((.((((((((((((..(((((......)))))(.(((....))).).....((....)))).))))))).)))(((.........))).)))))--------..------... ( -34.30) >DroVir_CAF1 170 105 - 1 ACACCAGCCAAAUUCGAUAGCAACGUCGACUUGCCCACCGAGGGGAAGCCGACGAAGCCCACCCGCGCAUCACCCGUCUUGGCCACCUCAAAGCCGGCUGC--------AA------AU- ....(((((......(((.((..(((((.(((.(((.....))).))).)))))..((......))))))).........(((.........)))))))).--------..------..- ( -34.50) >DroGri_CAF1 173 112 - 1 ACGCCGGCCAAGUUCGAGAGUAACGUCGACUUGCCCACCGAAGGAAAGCCGACGAAACCCACCCGGGCAUCACCAGUUUUGGCCACCUCAAAGCCGGCUAA--------GAGGAAAAACG .....((((((((((....).)))((((.(((..((......)).))).))))((..(((....)))..)).......)))))).((((..((....))..--------))))....... ( -33.60) >DroWil_CAF1 167 114 - 1 ACGCCAGCCAAAUUUGACAAUAACGUGGACUUCCCUACAGAGGGAAACCCUACAAAACCCACUCGAGCAUCUCCAGUUUUGGCCACCUCAAAGCCGGCUGCAAGAGAAGAA------AUA ..((.((((...(((((.......((((..((((((....))))))...))))....((.(((.(((...))).)))...)).....)))))...))))))..........------... ( -26.80) >DroMoj_CAF1 170 106 - 1 ACGCCAGCUAAAUUCGAUAGUAACGUCGAUUUACCCACAGAGGGAAAGCCGACGAAACCCACUCGCGCAUCUCCCGUCUUGGCCACCUCAAAGCCGGCUGA--------AA------AUG ..(((.(((....(((((......))))).....(((.(((((((..(((((..........))).))...)))).)))))).........))).)))...--------..------... ( -26.40) >DroAna_CAF1 172 109 - 1 ACGCCAGCUAAGUUGGAGAGGAGAGUGGACUUCCCCACUGACGGGAAGCCCACAAAGCCGACCCGGGCAUCUCCAGUCUUGGCCACCUCAAAGCCGGCUGAAUG-----AA------AAA ....(((((((((((((((.....((((.((((((.......)))))).))))...(((......))).))))))).)))(((.........))))))))....-----..------... ( -41.90) >consensus ACGCCAGCCAAAUUCGAGAGUAACGUCGACUUACCCACAGAGGGAAAGCCGACGAAACCCACCCGGGCAUCUCCAGUCUUGGCCACCUCAAAGCCGGCUGA________AA______AUA ....(((((...............((((.(((.((.......)).))).))))...........................(((.........)))))))).................... (-18.41 = -18.13 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:40 2006