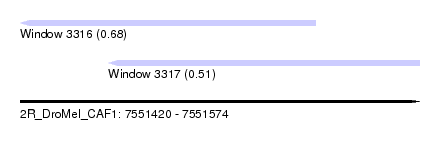
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,551,420 – 7,551,574 |
| Length | 154 |
| Max. P | 0.680614 |

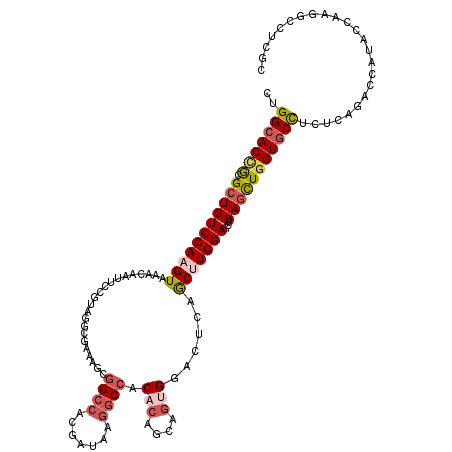
| Location | 7,551,420 – 7,551,534 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.81 |
| Mean single sequence MFE | -43.85 |
| Consensus MFE | -20.15 |
| Energy contribution | -19.60 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7551420 114 - 20766785 AGCUGCCACUAGAAGGCCACACAGCAGGGGACUCAGCCUGGAGCCCAGCUGGUGCCUGGUGGCCCACACCAAGGUCUCGUUCCACCUACUCUGCACCUGGUGGCUCAGCACCUG .(((((((((((.((((.((.((((..(((.(((......)))))).))))))))))(((((..(..((....))...)..)))))..........))))))))..)))..... ( -47.70) >DroVir_CAF1 9595 114 - 1 AACGGACACAAUAAGACCCCACAGCAGUGGGCUCAGCUUGGAGCGCAUUUGGUGCUGCUCCGACCAGUCCAAGGCAGCGCUCAGCUGUGCCUGCACAUAGCUGCUCAGAUGCUG ...((((.......((.(((((....))))).)).(.(((((((((((...)))).))))))).).))))....(((((((((((((((......))))))))...)).))))) ( -47.40) >DroPse_CAF1 11545 114 - 1 UGUGGCCACGAUGAGGCCACACAACAGUGGGCUCAACUUGGAGACCAGUUGGUGCCUCUCAGACCAUCCCAAGGCCUCGCUCCAUUGACCCUGUACAAAGCCGGUCAACUGUUG (((((((.......)))))))(((((((((((((((((.(....).)))))).))))...............((((..(((...(((........)))))).)))).))))))) ( -41.50) >DroEre_CAF1 12665 114 - 1 UGCGGCCACGAGAAGGCCACACAGGAGGGGACUCAGCCUGGAGCCCAGCUGGUGCCUGUUGGCCCACACCAAGGUCUCGUUCCACCGACUCUGCACCUGGUGGCUCAACUCCUG ((.((((.......)))).))(((((((((.(((......))))))((((((.(((....))))))(((((.((((..(((.....)))...).)))))))))))...)))))) ( -46.60) >DroAna_CAF1 9238 114 - 1 AAUGGCCACUAAGAAGCCACACAGAAAUGGACUCAGCUUGGAGGCCAGCUGGUGCCGUGUUGACCAUUCCAGGGAAGCGUUCCAAUGGCUCUGCACCUGUUGGUGCCACUCCUG ..(((((.(((((.(((((........))).))...))))).)))))..(((..((((((.(((((((...((((....))))))))).)).)))).....))..)))...... ( -38.40) >DroPer_CAF1 11509 114 - 1 UGUGGCCACGAUGAGGCCACACAACAGUGGGCUCAACUUGGAGACCAGUUGGUGCCUCUCAGACCAUCCCAAGGCCUCGCUCCAUUGACCCUGUACAAAGCCGGUCAACUGUUG (((((((.......)))))))(((((((((((((((((.(....).)))))).))))...............((((..(((...(((........)))))).)))).))))))) ( -41.50) >consensus AGCGGCCACGAUAAGGCCACACAGCAGUGGACUCAGCUUGGAGCCCAGCUGGUGCCUCUCAGACCAUACCAAGGCCUCGCUCCACUGACCCUGCACAUAGCGGCUCAACUCCUG ...((((.......))))...((((((.((((.((((.((.....)))))))).))......................(..(((((............)))))..)..)))))) (-20.15 = -19.60 + -0.55)

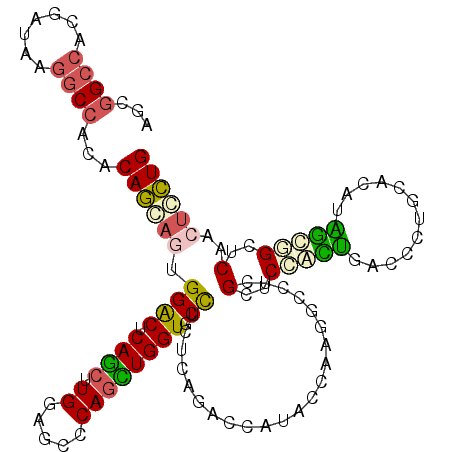
| Location | 7,551,454 – 7,551,574 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -44.28 |
| Consensus MFE | -25.85 |
| Energy contribution | -26.77 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7551454 120 - 20766785 CGGGCACCGCGCUGUCCAAGUACACGAUUCCGUACGCGAAAGCUGCCACUAGAAGGCCACACAGCAGGGGACUCAGCCUGGAGCCCAGCUGGUGCCUGGUGGCCCACACCAAGGUCUCGU (((((((((((((((....((((........))))((....)).(((.......)))...)))))..(((.(((......)))))).)).))))))))(.((((........)))).).. ( -49.30) >DroVir_CAF1 9629 120 - 1 CUGGCACAGCGCUGUCCAAGUAAACAAUGGCAUAGCAAAAAACGGACACAAUAAGACCCCACAGCAGUGGGCUCAGCUUGGAGCGCAUUUGGUGCUGCUCCGACCAGUCCAAGGCAGCGC ........((((((((...((........))............((((.......((.(((((....))))).)).(.(((((((((((...)))).))))))).).))))..)))))))) ( -44.60) >DroPse_CAF1 11579 120 - 1 CUGGCACUGCGCUGUCCAAGUAAACCAUUCCGUAGGCGAAUGUGGCCACGAUGAGGCCACACAACAGUGGGCUCAACUUGGAGACCAGUUGGUGCCUCUCAGACCAUCCCAAGGCCUCGC ..(((((((.(((((((((((.........(((.(((.......)))))).((((.((((......)))).)))))))))))...)))))))))))....((.((.......)).))... ( -42.80) >DroEre_CAF1 12699 120 - 1 CGGGCACCGCGCUGUCCAAGUACACGAUUCCGUAUGCGAAUGCGGCCACGAGAAGGCCACACAGGAGGGGACUCAGCCUGGAGCCCAGCUGGUGCCUGUUGGCCCACACCAAGGUCUCGU (((((((((.((((((((.(((.(((....))).)))....((((((.......))))......(((....))).)).))))...)))))))))))))..((((........)))).... ( -48.80) >DroAna_CAF1 9272 120 - 1 CCGGAACCACGCUGUCCAAGUAAACAAUUCCGUAGGCAAAAAUGGCCACUAAGAAGCCACACAGAAAUGGACUCAGCUUGGAGGCCAGCUGGUGCCGUGUUGACCAUUCCAGGGAAGCGU .(((.((((.(((((((((((..........((.(((.......)))))...((..(((........)))..)).)))))))...)))))))).))).(((..((......))..))).. ( -37.40) >DroPer_CAF1 11543 120 - 1 CUGGCACUGCGCUGUCCAAGUAAACCAUUCCGUAGGCGAAUGUGGCCACGAUGAGGCCACACAACAGUGGGCUCAACUUGGAGACCAGUUGGUGCCUCUCAGACCAUCCCAAGGCCUCGC ..(((((((.(((((((((((.........(((.(((.......)))))).((((.((((......)))).)))))))))))...)))))))))))....((.((.......)).))... ( -42.80) >consensus CUGGCACCGCGCUGUCCAAGUAAACAAUUCCGUAGGCGAAAGCGGCCACGAUAAGGCCACACAGCAGUGGACUCAGCUUGGAGCCCAGCUGGUGCCUCUCAGACCAUACCAAGGCCUCGC ..(((((((.(((((((((((......................((((.......)))).(((....)))......)))))))...)))))))))))........................ (-25.85 = -26.77 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:38 2006