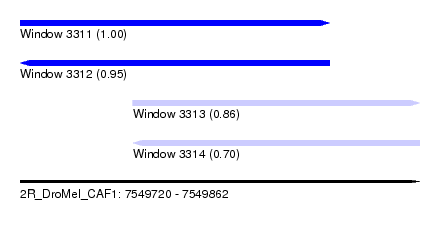
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 7,549,720 – 7,549,862 |
| Length | 142 |
| Max. P | 0.999592 |

| Location | 7,549,720 – 7,549,830 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.63 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -16.41 |
| Energy contribution | -16.47 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.68 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7549720 110 + 20766785 UUUGUUUCUAACUUUUUUCUUUUCAAUUUCAAAGUCGCUCUUAGGAGGGCUAAAACUAAUUU----------GAAUAUUGAUAACAAAUGAACUAUCGAUAAGAGGCCGUCGAUAGUUCA ((((((................((((((((((((((.(((....))))))).........))----------))).))))).))))))((((((((((((........)))))))))))) ( -26.63) >DroSim_CAF1 8257 109 + 1 UUUGUUUCAAACUUUUUCAU-UUCAAUUUUAAAGUCGCUCUUAGAAGGGCUAAAACAAAUUU----------GAAUAUAGAUAACAAAAGGGCUAUCGAUAAGAGACCGUCGAUAGUUCA ((((((((..(((((.....-.........))))).(((((....)))))............----------.......)).)))))).(((((((((((........))))))))))). ( -23.54) >DroEre_CAF1 10913 103 + 1 -----------------GCUUUUCAAUUUUAAAGUGAUUCAUAUAAGGGUUAAAACAAAUUUGAGUCCAGCUGAAUUCUGGUAACCAAUGAACUAUCGAUAAGAGGUCGUCGAUAGUUCA -----------------(((((........)))))............(((((...((...((.((.....)).))...)).)))))..((((((((((((........)))))))))))) ( -24.60) >DroYak_CAF1 8027 91 + 1 -----------------UGUCUUCAAUUUUAAAGUGGAUAAUAUAAGGGUUAAAUCAAA------------UGAAUUCUGAUAGCCAAUGAACUAUCGAUAAAAGGCUGUCGAUAGUUCA -----------------(((((.............))))).......(((((..(((..------------.......))))))))..(((((((((((((......))))))))))))) ( -22.02) >consensus _________________UCUUUUCAAUUUUAAAGUCGCUCAUAGAAGGGCUAAAACAAAUUU__________GAAUACUGAUAACAAAUGAACUAUCGAUAAGAGGCCGUCGAUAGUUCA ..................................((((((......)))))).....................................(((((((((((........))))))))))). (-16.41 = -16.47 + 0.06)

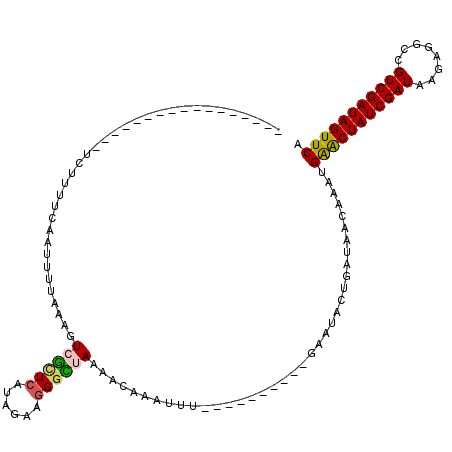
| Location | 7,549,720 – 7,549,830 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.63 |
| Mean single sequence MFE | -20.21 |
| Consensus MFE | -11.44 |
| Energy contribution | -11.75 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7549720 110 - 20766785 UGAACUAUCGACGGCCUCUUAUCGAUAGUUCAUUUGUUAUCAAUAUUC----------AAAUUAGUUUUAGCCCUCCUAAGAGCGACUUUGAAAUUGAAAAGAAAAAAGUUAGAAACAAA (((((((((((..(....)..)))))))))))(((.((.(((((.(((----------(((...(((((((.....)).)))))...))))))))))).)).)))............... ( -25.20) >DroSim_CAF1 8257 109 - 1 UGAACUAUCGACGGUCUCUUAUCGAUAGCCCUUUUGUUAUCUAUAUUC----------AAAUUUGUUUUAGCCCUUCUAAGAGCGACUUUAAAAUUGAA-AUGAAAAAGUUUGAAACAAA ....(((((((..(....)..)))))))....................----------...(((((((((((.((....)).))((((((.........-.....))))))))))))))) ( -16.24) >DroEre_CAF1 10913 103 - 1 UGAACUAUCGACGACCUCUUAUCGAUAGUUCAUUGGUUACCAGAAUUCAGCUGGACUCAAAUUUGUUUUAACCCUUAUAUGAAUCACUUUAAAAUUGAAAAGC----------------- (((((((((((..(....)..)))))))))))..(((((((((.......))))((........))..)))))..............................----------------- ( -21.50) >DroYak_CAF1 8027 91 - 1 UGAACUAUCGACAGCCUUUUAUCGAUAGUUCAUUGGCUAUCAGAAUUCA------------UUUGAUUUAACCCUUAUAUUAUCCACUUUAAAAUUGAAGACA----------------- (((((((((((..........)))))))))))..((..((((((.....------------))))))....)).............(((((....)))))...----------------- ( -17.90) >consensus UGAACUAUCGACGGCCUCUUAUCGAUAGUUCAUUGGUUAUCAAAAUUC__________AAAUUUGUUUUAACCCUUAUAAGAGCCACUUUAAAAUUGAAAAGA_________________ .((((((((((..(....)..))))))))))......................................................................................... (-11.44 = -11.75 + 0.31)

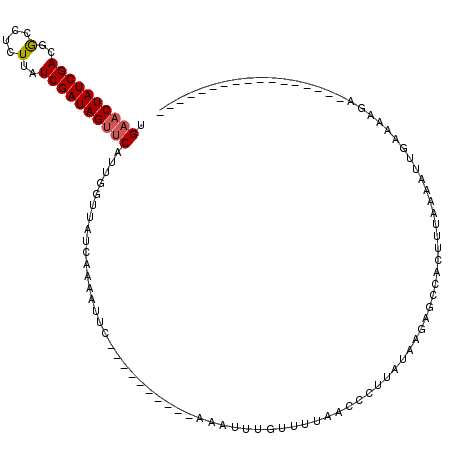
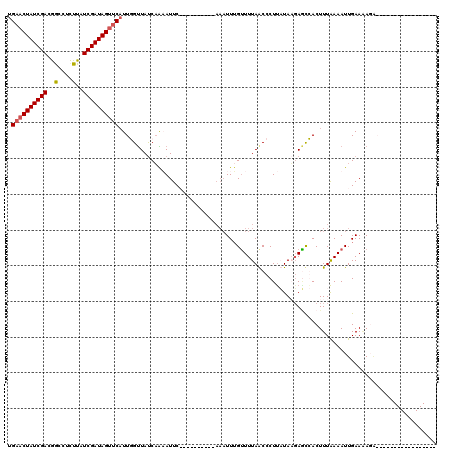
| Location | 7,549,760 – 7,549,862 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.19 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -17.60 |
| Energy contribution | -16.64 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7549760 102 + 20766785 UUAGGAGGGCUAAAACUAAUUU----------GAAUAUUGAUAACAAAUGAACUAUCGAUAAGAGGCCGUCGAUAGUUCACGGUGCAACAUCAACUCUCCCAUCACUAGUUC ...((((((...........((----------((...(((...((...((((((((((((........))))))))))))..)).)))..))))))))))............ ( -26.10) >DroSec_CAF1 8035 102 + 1 UUAGAAGGGCUAAAGCAAAUUU----------GAAUAUUGAUAACAAAUGUGCUAUCGAUAAGAGGCCGUCGAUAGUUCGCAGUACAACAUCAACUCUCCCAUCACUAGUUA ...((.((((....))......----------.....(((((......((((((((((((........)))))))))..))).......))))).....)).))........ ( -19.02) >DroSim_CAF1 8296 102 + 1 UUAGAAGGGCUAAAACAAAUUU----------GAAUAUAGAUAACAAAAGGGCUAUCGAUAAGAGACCGUCGAUAGUUCACAGUGCAACAUCAACUCUCCCAUCACUAGUUC ...((.(((.............----------.................(((((((((((........)))))))))))..(((.........)))..))).))........ ( -19.10) >DroEre_CAF1 10936 112 + 1 AUAUAAGGGUUAAAACAAAUUUGAGUCCAGCUGAAUUCUGGUAACCAAUGAACUAUCGAUAAGAGGUCGUCGAUAGUUCAGGAUGCAACAUUAAUUCUUCCAUCACUAGUUC ....(((((((((..((...((.((.....)).))...))(((.((..((((((((((((........)))))))))))))).)))....)))))))))............. ( -26.00) >DroYak_CAF1 8050 100 + 1 AUAUAAGGGUUAAAUCAAA------------UGAAUUCUGAUAGCCAAUGAACUAUCGAUAAAAGGCUGUCGAUAGUUCAGGGUGCAACAUCAAUUGUCCCACCACUAGUUC .......(((((..(((..------------.......))))))))..(((((((((((((......))))))))))))).((((..(((.....)))..))))........ ( -29.30) >consensus UUAGAAGGGCUAAAACAAAUUU__________GAAUAUUGAUAACAAAUGAACUAUCGAUAAGAGGCCGUCGAUAGUUCACGGUGCAACAUCAACUCUCCCAUCACUAGUUC ......(((........................................(((((((((((........)))))))))))..(((.........)))..)))........... (-17.60 = -16.64 + -0.96)

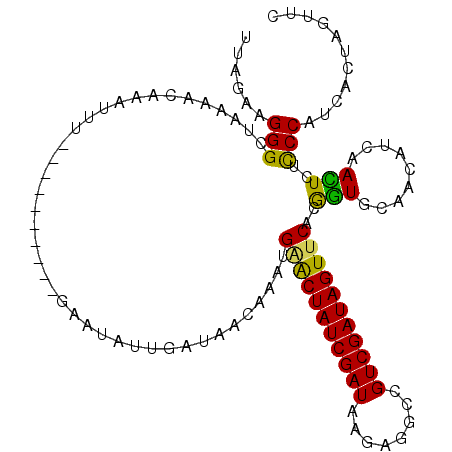
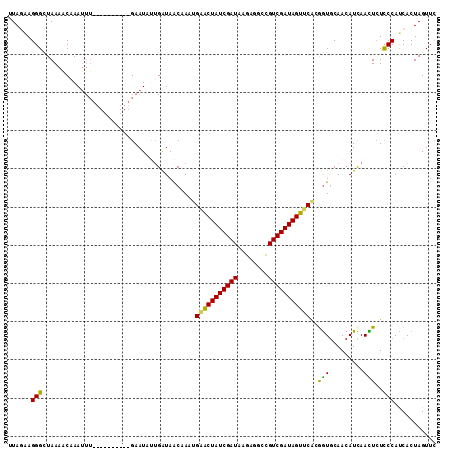
| Location | 7,549,760 – 7,549,862 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.19 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -16.36 |
| Energy contribution | -18.08 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 7549760 102 - 20766785 GAACUAGUGAUGGGAGAGUUGAUGUUGCACCGUGAACUAUCGACGGCCUCUUAUCGAUAGUUCAUUUGUUAUCAAUAUUC----------AAAUUAGUUUUAGCCCUCCUAA ((((((((.....(((.(((((((..(((..((((((((((((..(....)..)))))))))))).)))))))))).)))----------..))))))))............ ( -30.10) >DroSec_CAF1 8035 102 - 1 UAACUAGUGAUGGGAGAGUUGAUGUUGUACUGCGAACUAUCGACGGCCUCUUAUCGAUAGCACAUUUGUUAUCAAUAUUC----------AAAUUUGCUUUAGCCCUUCUAA ...((((.((((((((.((((((((((.....)))..)))))))...))))))))(((((((....))))))).......----------.........))))......... ( -23.80) >DroSim_CAF1 8296 102 - 1 GAACUAGUGAUGGGAGAGUUGAUGUUGCACUGUGAACUAUCGACGGUCUCUUAUCGAUAGCCCUUUUGUUAUCUAUAUUC----------AAAUUUGUUUUAGCCCUUCUAA ((((.(((((((((((((((((((((........))).))))))..)))))))))((((((......)))))).......----------..))).))))............ ( -22.90) >DroEre_CAF1 10936 112 - 1 GAACUAGUGAUGGAAGAAUUAAUGUUGCAUCCUGAACUAUCGACGACCUCUUAUCGAUAGUUCAUUGGUUACCAGAAUUCAGCUGGACUCAAAUUUGUUUUAACCCUUAUAU .(((((((((((.((.(.....).)).))))..((((((((((..(....)..))))))))))))))))).((((.......)))).......................... ( -26.40) >DroYak_CAF1 8050 100 - 1 GAACUAGUGGUGGGACAAUUGAUGUUGCACCCUGAACUAUCGACAGCCUUUUAUCGAUAGUUCAUUGGCUAUCAGAAUUCA------------UUUGAUUUAACCCUUAUAU .....((.((((.((((.....)))).))))))((((((((((..........))))))))))...((..((((((.....------------))))))....))....... ( -26.70) >consensus GAACUAGUGAUGGGAGAGUUGAUGUUGCACCGUGAACUAUCGACGGCCUCUUAUCGAUAGUUCAUUUGUUAUCAAUAUUC__________AAAUUUGUUUUAGCCCUUCUAA ((((((.(((((((((.(((((((((........))).))))))...))))))))).))))))................................................. (-16.36 = -18.08 + 1.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:36 2006